1G65
 
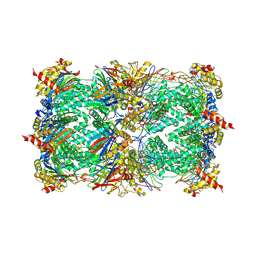 | | Crystal structure of epoxomicin:20s proteasome reveals a molecular basis for selectivity of alpha,beta-epoxyketone proteasome inhibitors | | Descriptor: | EPOXOMICIN (peptide inhibitor), MAGNESIUM ION, Proteasome component C1, ... | | Authors: | Groll, M, Kim, K.B, Kairies, N, Huber, R, Crews, C. | | Deposit date: | 2000-11-03 | | Release date: | 2000-11-27 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal Structure of Epoxomicin:20S Proteasome reveals a molecular basis for selectivity of alpha,beta-Epoxyketone Proteasome Inhibitors
J.Am.Chem.Soc., 122, 2000
|
|
1G3L
 
 | | THE STRUCTURAL BASIS OF THE CATALYTIC MECHANISM AND REGULATION OF GLUCOSE-1-PHOSPHATE THYMIDYLYLTRANSFERASE (RMLA). TDP-L-RHAMNOSE COMPLEX. | | Descriptor: | 2'-DEOXY-THYMIDINE-BETA-L-RHAMNOSE, GLUCOSE-1-PHOSPHATE THYMIDYLYLTRANSFERASE, SULFATE ION | | Authors: | Blankenfeldt, W, Asuncion, M, Lam, J.S, Naismith, J.H. | | Deposit date: | 2000-10-24 | | Release date: | 2000-12-27 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The structural basis of the catalytic mechanism and regulation of glucose-1-phosphate thymidylyltransferase (RmlA).
EMBO J., 19, 2000
|
|
1HQ1
 
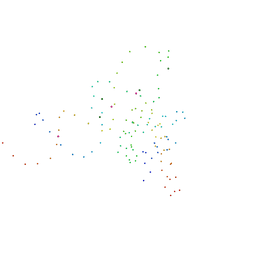 | | STRUCTURAL AND ENERGETIC ANALYSIS OF RNA RECOGNITION BY A UNIVERSALLY CONSERVED PROTEIN FROM THE SIGNAL RECOGNITION PARTICLE | | Descriptor: | 4.5S RNA DOMAIN IV, MAGNESIUM ION, POTASSIUM ION, ... | | Authors: | Batey, R.T, Sagar, M.B, Doudna, J.A. | | Deposit date: | 2000-12-13 | | Release date: | 2001-01-03 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Structural and energetic analysis of RNA recognition by a universally conserved protein from the signal recognition particle.
J.Mol.Biol., 307, 2001
|
|
1HWP
 
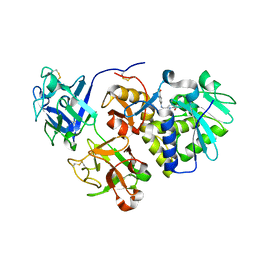 | | EBULIN COMPLEXED WITH PTEROIC ACID, TRIGONAL CRYSTAL FORM | | Descriptor: | EBULIN, PTEROIC ACID, beta-D-galactopyranose-(1-4)-beta-D-glucopyranose, ... | | Authors: | Pascal, J.M, Day, P.J, Monzingo, A.F, Ernst, S.R, Robertus, J.D. | | Deposit date: | 2001-01-09 | | Release date: | 2001-01-24 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | 2.8-A crystal structure of a nontoxic type-II ribosome-inactivating protein, ebulin l.
Proteins, 43, 2001
|
|
1HWY
 
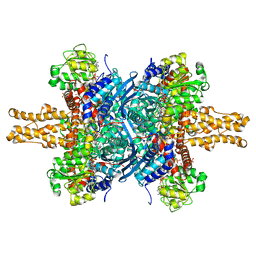 | | BOVINE GLUTAMATE DEHYDROGENASE COMPLEXED WITH NAD AND 2-OXOGLUTARATE | | Descriptor: | 2-OXOGLUTARIC ACID, GLUTAMATE DEHYDROGENASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Smith, T.J, Peterson, P.E, Schmidt, T, Fang, J, Stanley, C.A. | | Deposit date: | 2001-01-10 | | Release date: | 2001-01-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structures of bovine glutamate dehydrogenase complexes elucidate the mechanism of purine regulation.
J.Mol.Biol., 307, 2001
|
|
1HR0
 
 | | CRYSTAL STRUCTURE OF INITIATION FACTOR IF1 BOUND TO THE 30S RIBOSOMAL SUBUNIT | | Descriptor: | 16S RIBOSOMAL RNA, 30S RIBOSOMAL PROTEIN S10, 30S RIBOSOMAL PROTEIN S11, ... | | Authors: | Carter, A.P, Clemons Jr, W.M, Brodersen, D.E, Morgan-Warren, R.J, Wimberly, B.T, Ramakrishnan, V. | | Deposit date: | 2000-12-20 | | Release date: | 2001-01-24 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structure of an initiation factor bound to the 30S ribosomal subunit.
Science, 291, 2001
|
|
1HQ3
 
 | | CRYSTAL STRUCTURE OF THE HISTONE-CORE-OCTAMER IN KCL/PHOSPHATE | | Descriptor: | CHLORIDE ION, HISTONE H2A-IV, HISTONE H2B, ... | | Authors: | Chantalat, L, Nicholson, J.M, Lambert, S.J, Reid, A.J, Donovan, M.J, Reynolds, C.D, Wood, C.M, Baldwin, J.P. | | Deposit date: | 2000-12-14 | | Release date: | 2001-01-24 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structure of the histone-core octamer in KCl/phosphate crystals at 2.15 A resolution.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
1I2C
 
 | | CRYSTAL STRUCTURE OF MUTANT T145A SQD1 PROTEIN COMPLEX WITH NAD AND UDP-GLUCOSE | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SULFATE ION, SULFOLIPID BIOSYNTHESIS PROTEIN SQD1, ... | | Authors: | Theisen, M.J, Sanda, S.L, Ginell, S.L, Benning, C, Garavito, R.M. | | Deposit date: | 2001-02-07 | | Release date: | 2003-07-01 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Characterization of the Active Site of UDP-sulfoquinovose Synthase: Formation of the Sulfonic Acid Product in the Crystalline State.
To be Published
|
|
1I94
 
 | | CRYSTAL STRUCTURES OF THE SMALL RIBOSOMAL SUBUNIT WITH TETRACYCLINE, EDEINE AND IF3 | | Descriptor: | 16S RRNA, 30S RIBOSOMAL PROTEIN S10, 30S RIBOSOMAL PROTEIN S11, ... | | Authors: | Pioletti, M, Schluenzen, F, Harms, J, Zarivach, R, Gluehmann, M, Avila, H, Bartels, H, Jacobi, C, Hartsch, T, Yonath, A, Franceschi, F. | | Deposit date: | 2001-03-18 | | Release date: | 2001-04-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structures of complexes of the small ribosomal subunit with tetracycline, edeine and IF3.
EMBO J., 20, 2001
|
|
1FXO
 
 | |
1G1L
 
 | | THE STRUCTURAL BASIS OF THE CATALYTIC MECHANISM AND REGULATION OF GLUCOSE-1-PHOSPHATE THYMIDYLYLTRANSFERASE (RMLA). TDP-GLUCOSE COMPLEX. | | Descriptor: | 2'DEOXY-THYMIDINE-5'-DIPHOSPHO-ALPHA-D-GLUCOSE, CITRIC ACID, GLUCOSE-1-PHOSPHATE THYMIDYLYLTRANSFERASE, ... | | Authors: | Blankenfeldt, W, Asuncion, M, Lam, J.S, Naimsmith, J.H. | | Deposit date: | 2000-10-12 | | Release date: | 2000-12-27 | | Last modified: | 2018-01-31 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | The structural basis of the catalytic mechanism and regulation of glucose-1-phosphate thymidylyltransferase (RmlA).
EMBO J., 19, 2000
|
|
1G4G
 
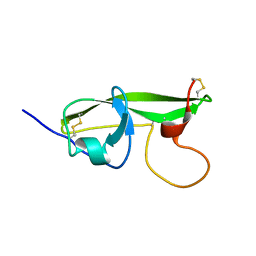 | | NMR STRUCTURE OF THE FIFTH DOMAIN OF HUMAN BETA2-GLYCOPROTEIN I | | Descriptor: | BETA2-GLYCOPROTEIN I | | Authors: | Hoshino, M, Hagihara, Y, Nishii, I, Yamazaki, T, Kato, H, Goto, Y. | | Deposit date: | 2000-10-27 | | Release date: | 2000-11-15 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Identification of the phospholipid-binding site of human beta(2)-glycoprotein I domain V by heteronuclear magnetic resonance.
J.Mol.Biol., 304, 2000
|
|
1JB0
 
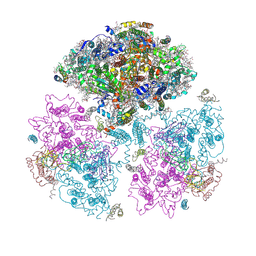 | | Crystal Structure of Photosystem I: a Photosynthetic Reaction Center and Core Antenna System from Cyanobacteria | | Descriptor: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, BETA-CAROTENE, ... | | Authors: | Jordan, P, Fromme, P, Witt, H.T, Klukas, O, Saenger, W, Krauss, N. | | Deposit date: | 2001-06-01 | | Release date: | 2001-08-01 | | Last modified: | 2019-11-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Three-dimensional Structure of Cyanobacterial Photosystem I at 2.5 A Resolution
NATURE, 411, 2001
|
|
1JQY
 
 | | HEAT-LABILE ENTEROTOXIN B-PENTAMER WITH LIGAND BMSC-0010 | | Descriptor: | (3-NITRO-5-(3-MORPHOLIN-4-YL-PROPYLAMINOCARBONYL)PHENYL)-GALACTOPYRANOSIDE, HEAT-LABILE ENTEROTOXIN B CHAIN | | Authors: | Merritt, E.A, Hol, W.G.J. | | Deposit date: | 2001-08-09 | | Release date: | 2002-05-08 | | Last modified: | 2017-10-04 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Anchor-based design of improved cholera toxin and E. coli heat-labile enterotoxin receptor binding antagonists that display multiple binding modes.
Chem.Biol., 9, 2002
|
|
1GCU
 
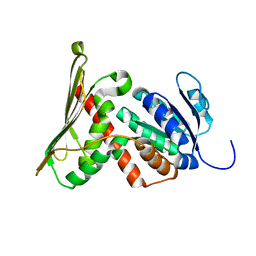 | |
1GEG
 
 | | CRYATAL STRUCTURE ANALYSIS OF MESO-2,3-BUTANEDIOL DEHYDROGENASE | | Descriptor: | ACETOIN REDUCTASE, BETA-MERCAPTOETHANOL, MAGNESIUM ION, ... | | Authors: | Otagiri, M, Kurisu, G, Ui, S, Kusunoki, M. | | Deposit date: | 2000-11-10 | | Release date: | 2001-02-28 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of meso-2,3-butanediol dehydrogenase in a complex with NAD+ and inhibitor mercaptoethanol at 1.7 A resolution for understanding of chiral substrate recognition mechanisms.
J.Biochem., 129, 2001
|
|
1JD2
 
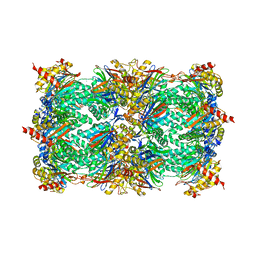 | | Crystal Structure of the yeast 20S Proteasome:TMC-95A complex: A non-covalent Proteasome Inhibitor | | Descriptor: | MAGNESIUM ION, PROTEASOME COMPONENT C1, PROTEASOME COMPONENT C11, ... | | Authors: | Groll, M, Koguchi, Y, Huber, R, Kohno, J. | | Deposit date: | 2001-06-12 | | Release date: | 2002-02-13 | | Last modified: | 2017-10-04 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of the 20 S proteasome:TMC-95A complex: a non-covalent proteasome inhibitor.
J.Mol.Biol., 311, 2001
|
|
1JQG
 
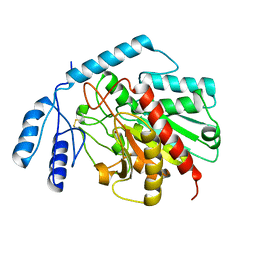 | | Crystal Structure of the Carboxypeptidase A from Helicoverpa Armigera | | Descriptor: | ZINC ION, carboxypeptidase A | | Authors: | Estebanez-Perpina, E, Bayes, A, Vendrell, J, Jongsma, M.A, Bown, D.P, Gatehouse, J.A, Huber, R, Bode, W, Aviles, F.X, Reverter, D. | | Deposit date: | 2001-08-07 | | Release date: | 2002-08-07 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of a novel mid-gut procarboxypeptidase from the cotton pest Helicoverpa armigera.
J.Mol.Biol., 313, 2001
|
|
1JRI
 
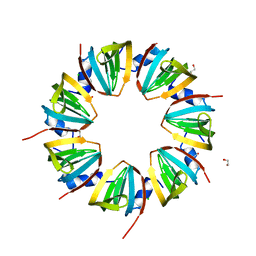 | |
1JO6
 
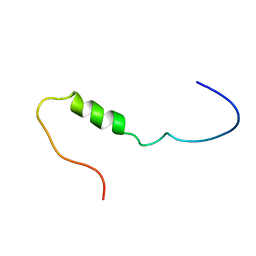 | | Solution structure of the cytoplasmic N-terminus of the BK beta-subunit KCNMB2 | | Descriptor: | potassium large conductance calcium-activated channel, subfamily M, beta member 2 | | Authors: | Bentrop, D, Beyermann, M, Wissmann, R, Fakler, B. | | Deposit date: | 2001-07-27 | | Release date: | 2001-11-16 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the "ball-and-chain" domain of KCNMB2, the beta 2-subunit of large conductance Ca2+- and voltage-activated potassium channels.
J.Biol.Chem., 276, 2001
|
|
1ILC
 
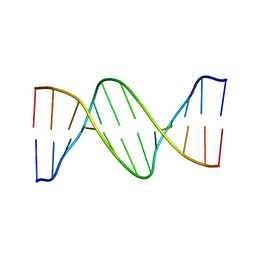 | | DNA Bending by an Adenine-Thymine Tract and Its Role in Gene Regulation. | | Descriptor: | 5'-D(*AP*CP*CP*GP*AP*AP*TP*TP*CP*GP*GP*T)-3' | | Authors: | Hizver, J, Rozenberg, H, Frolow, F, Rabinovich, D, Shakked, Z. | | Deposit date: | 2001-05-08 | | Release date: | 2002-05-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | DNA bending by an adenine--thymine tract and its role in gene regulation.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
1J0B
 
 | | Crystal Structure Analysis of the ACC deaminase homologue complexed with inhibitor | | Descriptor: | 1-aminocyclopropane-1-carboxylate deaminase, N-[3-HYDROXY-2-METHYL-5-PHOSPHONOOXYMETHYL-PYRIDIN-4-Y-LMETHYL]-1-AMINO-CYCLOPROPANECARBOXYLIC ACID | | Authors: | Fujino, A, Ose, T, Honma, M, Yao, M, Tanaka, I. | | Deposit date: | 2002-11-12 | | Release date: | 2003-05-12 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural and enzymatic properties of 1-aminocyclopropane-1-carboxylate deaminase homologue from Pyrococcus horikoshii
J.Mol.Biol., 341, 2004
|
|
1JI4
 
 | | NAP protein from helicobacter pylori | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, FE (III) ION, NEUTROPHIL-ACTIVATING PROTEIN A, ... | | Authors: | Zanotti, G, Papinutto, E, Dundon, W.G, Battistutta, R, Seveso, M, Del Giudice, G, Rappuoli, R, Montecucco, C. | | Deposit date: | 2001-06-29 | | Release date: | 2002-10-09 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Structure of the Neutrophil-activating Protein from Helicobacter pylori
J.Mol.Biol., 323, 2002
|
|
1IRH
 
 | | The Solution Structure of The Third Kunitz Domain of Tissue Factor Pathway Inhibitor | | Descriptor: | tissue factor pathway inhibitor | | Authors: | Mine, S, Yamazaki, T, Miyata, T, Hara, S, Kato, H. | | Deposit date: | 2001-10-02 | | Release date: | 2002-02-06 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Structural mechanism for heparin-binding of the third Kunitz domain of human tissue factor pathway inhibitor.
Biochemistry, 41, 2002
|
|
1IZL
 
 | | Crystal Structure of Photosystem II | | Descriptor: | 2-[(3-HYDROXY-2-METHYL-5-PHOSPHONOOXYMETHYL-PYRIDIN-4-YLMETHYL)-AMINO]-2-METHYL-SUCCINIC ACID, BETA-CAROTENE, CHLOROPHYLL A, ... | | Authors: | Kamiya, N, Shen, J.-R. | | Deposit date: | 2002-10-04 | | Release date: | 2003-01-14 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Crystal structure of oxygen-evolving photosystem II from Thermosynechococcus vulcanus at 3.7-A resolution
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
