1CUW
 
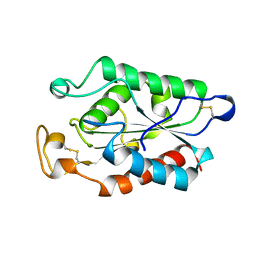 | | CUTINASE, G82A, A85F, V184I, A185L, L189F MUTANT | | Descriptor: | CUTINASE | | Authors: | Longhi, S, Cambillau, C. | | Deposit date: | 1995-11-16 | | Release date: | 1996-07-11 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Dynamics of Fusarium solani cutinase investigated through structural comparison among different crystal forms of its variants.
Proteins, 26, 1996
|
|
1CUX
 
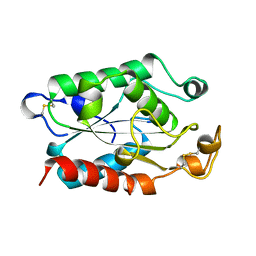 | | CUTINASE, L114Y MUTANT | | Descriptor: | CUTINASE | | Authors: | Longhi, S, Cambillau, C. | | Deposit date: | 1995-11-16 | | Release date: | 1996-07-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Dynamics of Fusarium solani cutinase investigated through structural comparison among different crystal forms of its variants.
Proteins, 26, 1996
|
|
1CUY
 
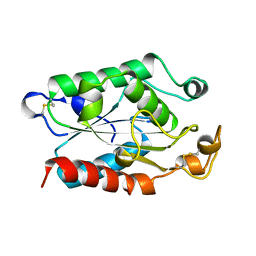 | | CUTINASE, L189F MUTANT | | Descriptor: | CUTINASE | | Authors: | Longhi, S, Cambillau, C. | | Deposit date: | 1995-11-16 | | Release date: | 1996-07-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Dynamics of Fusarium solani cutinase investigated through structural comparison among different crystal forms of its variants.
Proteins, 26, 1996
|
|
1CUZ
 
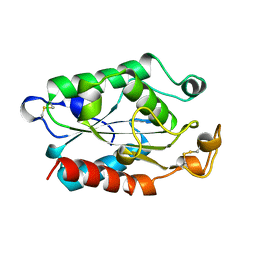 | | CUTINASE, L81G, L182G MUTANT | | Descriptor: | CUTINASE | | Authors: | Nicolas, A, Cambillau, C. | | Deposit date: | 1995-11-16 | | Release date: | 1996-07-11 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Dynamics of Fusarium solani cutinase investigated through structural comparison among different crystal forms of its variants.
Proteins, 26, 1996
|
|
1CV0
 
 | | T4 LYSOZYME MUTANT F104M | | Descriptor: | 2-HYDROXYETHYL DISULFIDE, CHLORIDE ION, LYSOZYME | | Authors: | Gassner, N.C, Baase, W.A, Lindstrom, J.D, Lu, J, Matthews, B.W. | | Deposit date: | 1999-08-20 | | Release date: | 1999-11-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Methionine and alanine substitutions show that the formation of wild-type-like structure in the carboxy-terminal domain of T4 lysozyme is a rate-limiting step in folding.
Biochemistry, 38, 1999
|
|
1CV1
 
 | | T4 LYSOZYME MUTANT V111M | | Descriptor: | 2-HYDROXYETHYL DISULFIDE, CHLORIDE ION, LYSOZYME | | Authors: | Gassner, N.C, Baase, W.A, Lindstrom, J.D, Lu, J, Matthews, B.W. | | Deposit date: | 1999-08-20 | | Release date: | 1999-11-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Methionine and alanine substitutions show that the formation of wild-type-like structure in the carboxy-terminal domain of T4 lysozyme is a rate-limiting step in folding.
Biochemistry, 38, 1999
|
|
1CV2
 
 | | Hydrolytic haloalkane dehalogenase linb from sphingomonas paucimobilis UT26 AT 1.6 A resolution | | Descriptor: | HALOALKANE DEHALOGENASE | | Authors: | Marek, J, Vevodova, J, Damborsky, J, Smatanova, I, Svensson, L.A, Newman, J, Nagata, Y, Takagi, M. | | Deposit date: | 1999-08-22 | | Release date: | 2000-09-11 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Crystal structure of the haloalkane dehalogenase from Sphingomonas paucimobilis UT26.
Biochemistry, 39, 2000
|
|
1CV3
 
 | | T4 LYSOZYME MUTANT L121M | | Descriptor: | 2-HYDROXYETHYL DISULFIDE, CHLORIDE ION, LYSOZYME | | Authors: | Gassner, N.C, Baase, W.A, Lindstrom, J, Lu, J, Matthews, B.W. | | Deposit date: | 1999-08-22 | | Release date: | 1999-08-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Methionine and alanine substitutions show that the formation of wild-type-like structure in the carboxy-terminal domain of T4 lysozyme is a rate-limiting step in folding.
Biochemistry, 38, 1999
|
|
1CV4
 
 | | T4 LYSOZYME MUTANT L118M | | Descriptor: | 2-HYDROXYETHYL DISULFIDE, CHLORIDE ION, LYSOZYME | | Authors: | Gassner, N.C, Baase, W.A, Lindstrom, J, Lu, J, Matthews, B.W. | | Deposit date: | 1999-08-22 | | Release date: | 1999-11-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Methionine and alanine substitutions show that the formation of wild-type-like structure in the carboxy-terminal domain of T4 lysozyme is a rate-limiting step in folding.
Biochemistry, 38, 1999
|
|
1CV5
 
 | | T4 LYSOZYME MUTANT L133M | | Descriptor: | 2-HYDROXYETHYL DISULFIDE, CHLORIDE ION, LYSOZYME | | Authors: | Gassner, N.C, Baase, W.A, Lindstrom, J, Lu, J, Matthews, B.W. | | Deposit date: | 1999-08-22 | | Release date: | 1999-11-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Methionine and alanine substitutions show that the formation of wild-type-like structure in the carboxy-terminal domain of T4 lysozyme is a rate-limiting step in folding.
Biochemistry, 38, 1999
|
|
1CV6
 
 | | T4 LYSOZYME MUTANT V149M | | Descriptor: | 2-HYDROXYETHYL DISULFIDE, CHLORIDE ION, LYSOZYME | | Authors: | Gassner, N.C, Baase, W.A, Lindstrom, J, Lu, J, Matthews, B.W. | | Deposit date: | 1999-08-22 | | Release date: | 1999-11-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Methionine and alanine substitutions show that the formation of wild-type-like structure in the carboxy-terminal domain of T4 lysozyme is a rate-limiting step in folding.
Biochemistry, 38, 1999
|
|
1CV7
 
 | |
1CV8
 
 | | STAPHOPAIN, CYSTEINE PROTEINASE FROM STAPHYLOCOCCUS AUREUS V8 | | Descriptor: | ACETATE ION, N-[N-[1-HYDROXYCARBOXYETHYL-CARBONYL]LEUCYLAMINO-BUTYL]-GUANIDINE, STAPHOPAIN | | Authors: | Hofmann, B, Schomburg, D, Hecht, H.-J. | | Deposit date: | 1998-05-08 | | Release date: | 1998-10-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal Structure of a Thiol Proteinase from Staphylococcus Aureus V-8 in the E-64 Inhibitor Complex
Acta Crystallogr.,Sect.A, 49, 1993
|
|
1CV9
 
 | | NMR STUDY OF ITAM PEPTIDE SUBSTRATE | | Descriptor: | IG-ALPHA ITAM PEPTIDE | | Authors: | Gaul, B.S, Harrison, M.L, Geahlen, R.L, Post, C.B. | | Deposit date: | 1999-08-23 | | Release date: | 1999-08-31 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Substrate recognition by the Lyn protein-tyrosine kinase. NMR structure of the immunoreceptor tyrosine-based activation motif signaling region of the B cell antigen receptor.
J.Biol.Chem., 275, 2000
|
|
1CVA
 
 | |
1CVB
 
 | |
1CVC
 
 | |
1CVD
 
 | |
1CVE
 
 | |
1CVF
 
 | |
1CVH
 
 | |
1CVI
 
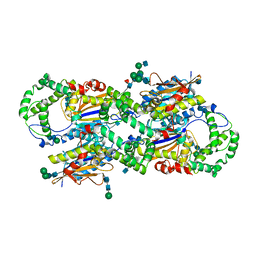 | | CRYSTAL STRUCTURE OF HUMAN PROSTATIC ACID PHOSPHATASE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCINE, ... | | Authors: | Jakob, C.G, Lewinski, K, Kuciel, R, Ostrowski, W, Lebioda, L. | | Deposit date: | 1999-08-23 | | Release date: | 1999-08-31 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structure of human prostatic acid phosphatase .
Prostate, 42, 2000
|
|
1CVJ
 
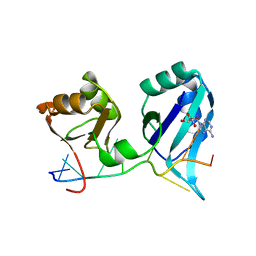 | | X-RAY CRYSTAL STRUCTURE OF THE POLY(A)-BINDING PROTEIN IN COMPLEX WITH POLYADENYLATE RNA | | Descriptor: | 5'-R(*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*A)-3', ADENOSINE MONOPHOSPHATE, POLYADENYLATE BINDING PROTEIN 1 | | Authors: | Deo, R.C, Bonanno, J.B, Sonenberg, N, Burley, S.K. | | Deposit date: | 1999-08-23 | | Release date: | 1999-10-04 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Recognition of polyadenylate RNA by the poly(A)-binding protein.
Cell(Cambridge,Mass.), 98, 1999
|
|
1CVK
 
 | | T4 LYSOZYME MUTANT L118A | | Descriptor: | 2-HYDROXYETHYL DISULFIDE, CHLORIDE ION, LYSOZYME | | Authors: | Gassner, N.C, Baase, W.A, Lindstrom, J, Lu, J, Matthews, B.W. | | Deposit date: | 1999-08-23 | | Release date: | 1999-11-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Methionine and alanine substitutions show that the formation of wild-type-like structure in the carboxy-terminal domain of T4 lysozyme is a rate-limiting step in folding.
Biochemistry, 38, 1999
|
|
1CVL
 
 | | CRYSTAL STRUCTURE OF BACTERIAL LIPASE FROM CHROMOBACTERIUM VISCOSUM ATCC 6918 | | Descriptor: | CALCIUM ION, TRIACYLGLYCEROL HYDROLASE | | Authors: | Lang, D.A, Hofmann, B, Haalck, L, Hecht, H.-J, Spener, F, Schmid, R.D, Schomburg, D. | | Deposit date: | 1997-01-09 | | Release date: | 1997-04-01 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of a bacterial lipase from Chromobacterium viscosum ATCC 6918 refined at 1.6 angstroms resolution.
J.Mol.Biol., 259, 1996
|
|
