7EOJ
 
 | |
2XNX
 
 | | BC1 fragment of streptococcal M1 protein in complex with human fibrinogen | | Descriptor: | FIBRINOGEN ALPHA CHAIN, FIBRINOGEN BETA CHAIN, FIBRINOGEN GAMMA CHAIN, ... | | Authors: | Macheboeuf, P, Y Fu, C, Zinkernagel, A.S, Johnson, J.E, Nizet, V, Ghosh, P. | | Deposit date: | 2010-08-06 | | Release date: | 2011-04-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Streptococcal M1 Protein Constructs a Pathological Host Fibrinogen Network
Nature, 472, 2011
|
|
7EOG
 
 | |
7QQN
 
 | | The PDZ domain of SNTG1 complexed with the acetylated PDZ-binding motif of TRPV3 | | Descriptor: | CALCIUM ION, GLYCEROL, Gamma-1-syntrophin,Annexin A2, ... | | Authors: | Cousido-Siah, A, Trave, G, Gogl, G. | | Deposit date: | 2022-01-10 | | Release date: | 2022-04-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | A scalable strategy to solve structures of PDZ domains and their complexes.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7EOI
 
 | |
5LBB
 
 | |
7AP7
 
 | | Structure of the W64R amyloidogenic variant of human lysozyme | | Descriptor: | Lysozyme C, SULFATE ION | | Authors: | Vettore, N, Herman, R, Kerff, F, Charlier, P, Sauvage, E, Brans, A, Morray, J, Dobson, C, Kumita, J, Dumoulin, M. | | Deposit date: | 2020-10-16 | | Release date: | 2021-08-25 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Characterisation of the structural, dynamic and aggregation properties of the W64R amyloidogenic variant of human lysozyme.
Biophys.Chem., 271, 2021
|
|
5LBN
 
 | |
2XPY
 
 | | Structure of Native Leukotriene A4 Hydrolase from Saccharomyces cerevisiae | | Descriptor: | GLUTATHIONE, GLYCEROL, LEUKOTRIENE A-4 HYDROLASE, ... | | Authors: | Helgstrand, C, Hasan, M, Usyal, H, Haeggstrom, J.Z, Thunnissen, M.M.G.M. | | Deposit date: | 2010-08-31 | | Release date: | 2010-12-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | A Leukotriene A(4) Hydrolase-Related Aminopeptidase from Yeast Undergoes Induced Fit Upon Inhibitor Binding.
J.Mol.Biol., 406, 2011
|
|
2M33
 
 | | Solution NMR structure of full-length oxidized microsomal rabbit cytochrome b5 | | Descriptor: | Cytochrome b5, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Subramanian, V, Ahuja, S, Popovych, N, Huang, R, Le Clair, S.V, Jahr, N, Soong, R, Xu, J, Yamamoto, K, Nanga, R.P, Im, S, Waskell, L, Ramamoorthy, A. | | Deposit date: | 2013-01-08 | | Release date: | 2013-02-06 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR structure of full-length mammalian cytochrome b5
To be Published
|
|
7KQV
 
 | |
7FIE
 
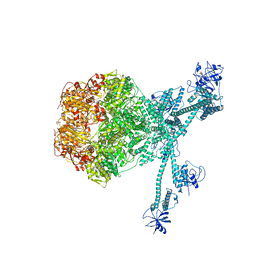 | | Processive cleavage of substrate at individual proteolytic active sites of the Lon protease complex (conformation 2) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Lon protease, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, ... | | Authors: | Li, S, Hsieh, K, Kuo, C, Su, S, Huang, K, Zhang, K, Chang, C.I. | | Deposit date: | 2021-07-31 | | Release date: | 2021-11-24 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.36 Å) | | Cite: | Processive cleavage of substrate at individual proteolytic active sites of the Lon protease complex.
Sci Adv, 7, 2021
|
|
3II0
 
 | | Crystal structure of human Glutamate oxaloacetate transaminase 1 (GOT1) | | Descriptor: | 1,2-ETHANEDIOL, Aspartate aminotransferase, cytoplasmic, ... | | Authors: | Ugochukwu, E, Pilka, E, Cooper, C, Bray, J.E, Yue, W.W, Muniz, J, Chaikuad, A, von Delft, F, Bountra, C, Arrowsmith, C.H, Weigelt, J, Edwards, A, Kavanagh, K.L, Oppermann, U, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-07-31 | | Release date: | 2009-08-11 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structure of human Glutamate oxaloacetate transaminase 1 (GOT1)
To be Published
|
|
2XTZ
 
 | | Crystal structure of the G alpha protein AtGPA1 from Arabidopsis thaliana | | Descriptor: | 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE, CHLORIDE ION, GUANINE NUCLEOTIDE-BINDING PROTEIN ALPHA-1 SUBUNIT, ... | | Authors: | Jones, J.C, Duffy, J.W, Machius, M, Temple, B.R.S, Dohlman, H.G, Jones, A.M. | | Deposit date: | 2010-10-13 | | Release date: | 2011-02-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | The Crystal Structure of a Self-Activating G Protein Alpha Subunit Reveals its Distinct Mechanism of Signal Initiation
Sci.Signal., 159, 2011
|
|
7EON
 
 | | Crystal structure of the Pepper aptamer in complex with HBC514 | | Descriptor: | 4-[(~{Z})-1-cyano-2-[4-[2-(dimethylamino)ethyl-methyl-amino]phenyl]ethenyl]benzenecarbonitrile, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Huang, K.Y, Ren, A.M. | | Deposit date: | 2021-04-22 | | Release date: | 2021-11-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure-based investigation of fluorogenic Pepper aptamer.
Nat.Chem.Biol., 17, 2021
|
|
2M0R
 
 | | Solution structure and dynamics of human S100A14 | | Descriptor: | Protein S100-A14 | | Authors: | Bertini, I, Borsi, V, Cerofolini, L, Das Gupta, S, Fragai, M, Luchinat, C. | | Deposit date: | 2012-11-05 | | Release date: | 2013-01-23 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure and dynamics of human S100A14.
J.Biol.Inorg.Chem., 18, 2013
|
|
7T9U
 
 | | Crystal structure of hSTING with an agonist (SHR169224) | | Descriptor: | (3S,4S)-2-[(1-ethyl-3-methyl-1H-pyrazole-5-carbonyl)amino]-4-(prop-2-en-1-yl)-4,5-dihydroimidazo[1,5,4-de][1,4]benzoxazine-8-carboxamide, CALCIUM ION, Stimulator of interferon genes protein | | Authors: | Chowdhury, R, Miller, M. | | Deposit date: | 2021-12-20 | | Release date: | 2022-12-21 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | SHR1032, a novel STING agonist, stimulates anti-tumor immunity and directly induces AML apoptosis.
Sci Rep, 12, 2022
|
|
2XQV
 
 | | The X-ray structure of the Zn(II) bound ZnuA from Salmonella enterica | | Descriptor: | SULFATE ION, ZINC ABC TRANSPORTER, PERIPLASMIC ZINC-BINDING PROTEIN, ... | | Authors: | Alaleona, F, Ilari, A, Chiancone, E, Battistoni, A, Petrarca, P. | | Deposit date: | 2010-09-07 | | Release date: | 2011-04-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | The X-Ray Structure of the Zinc Transporter Znua from Salmonella Enterica Discloses a Unique Triad of Zinc Coordinating Histidines.
J.Mol.Biol., 409, 2011
|
|
2XSZ
 
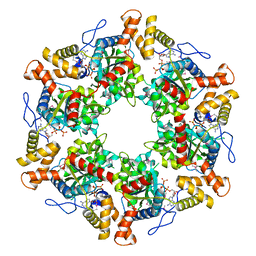 | | The dodecameric human RuvBL1:RuvBL2 complex with truncated domains II | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, RUVB-LIKE 1, RUVB-LIKE 2 | | Authors: | Gorynia, S, Bandeiras, T.M, Matias, P.M, Pinho, F.G, McVey, C.E, Vonrhein, C, Svergun, D.I, Round, A, Donner, P, Carrondo, M.A. | | Deposit date: | 2010-10-01 | | Release date: | 2011-10-05 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural and Functional Insights Into a Dodecameric Molecular Machine - the Ruvbl1/Ruvbl2 Complex.
J.Struct.Biol., 176, 2011
|
|
7EOM
 
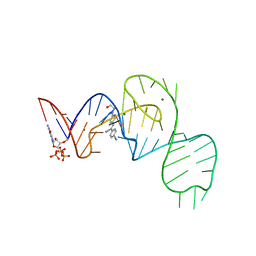 | | Crystal structure of the Pepper aptamer in complex with HBC508 | | Descriptor: | 4-[(~{Z})-1-cyano-2-[6-[2-hydroxyethyl(methyl)amino]pyridin-3-yl]ethenyl]benzenecarbonitrile, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Huang, K.Y, Ren, A.M. | | Deposit date: | 2021-04-22 | | Release date: | 2021-11-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.703 Å) | | Cite: | Structure-based investigation of fluorogenic Pepper aptamer.
Nat.Chem.Biol., 17, 2021
|
|
7EOH
 
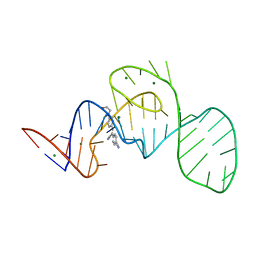 | | Crystal structure of the Pepper aptamer in complex with HBC | | Descriptor: | 4-[(~{Z})-1-cyano-2-[4-[2-hydroxyethyl(methyl)amino]phenyl]ethenyl]benzenecarbonitrile, MAGNESIUM ION, Pepper (49-MER) | | Authors: | Huang, K.Y, Ren, A.M. | | Deposit date: | 2021-04-22 | | Release date: | 2021-11-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.637 Å) | | Cite: | Structure-based investigation of fluorogenic Pepper aptamer.
Nat.Chem.Biol., 17, 2021
|
|
7B4J
 
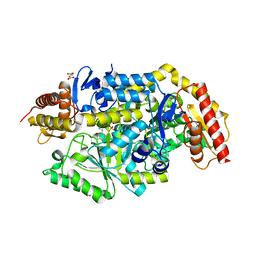 | | Thermostable omega transaminase PjTA-R6 variant W58M/F86L/R417L engineered for asymmetric synthesis of enantiopure bulky amines | | Descriptor: | 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, Aspartate aminotransferase family protein, SUCCINIC ACID | | Authors: | Capra, N, Rozeboom, H.J, Thunnissen, A.M.W.H, Janssen, D.B. | | Deposit date: | 2020-12-02 | | Release date: | 2021-09-01 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Computational Redesign of an omega-Transaminase from Pseudomonas jessenii for Asymmetric Synthesis of Enantiopure Bulky Amines.
Acs Catalysis, 11, 2021
|
|
5L8E
 
 | | Structure of UAF1 | | Descriptor: | GLYCEROL, Unknown, WD repeat-containing protein 48 | | Authors: | Dharadhar, S, Sixma, T. | | Deposit date: | 2016-06-07 | | Release date: | 2016-09-28 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A conserved two-step binding for the UAF1 regulator to the USP12 deubiquitinating enzyme.
J.Struct.Biol., 196, 2016
|
|
2Y1P
 
 | | Catalytic domain of mouse 2',3'-cyclic nucleotide 3'- phosphodiesterase, complexed with citrate | | Descriptor: | 2', 3'-CYCLIC NUCLEOTIDE 3'-PHOSPHODIESTERASE, CITRIC ACID, ... | | Authors: | Myllykoski, M, Kursula, P. | | Deposit date: | 2010-12-09 | | Release date: | 2011-12-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Myelin 2',3'-Cyclic Nucleotide 3'-Phosphodiesterase: Active-Site Ligand Binding and Molecular Conformation.
Plos One, 7, 2012
|
|
7AQS
 
 | | Crystal structure of E. coli DPS in space group P1 | | Descriptor: | DNA protection during starvation protein, FE (III) ION | | Authors: | Jakob, R.P, Pipercevic, J, Righetto, R, Goldie, K, Stahlberg, H, Maier, T, Hiller, S. | | Deposit date: | 2020-10-22 | | Release date: | 2021-09-01 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Identification of a Dps contamination in Mitomycin-C-induced expression of Colicin Ia.
Biochim Biophys Acta Biomembr, 1863, 2021
|
|
