1X9H
 
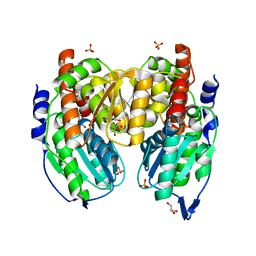 | | Crystal structure of phosphoglucose/phosphomannose isomerase from Pyrobaculum aerophilum in complex with fructose 6-phosphate | | Descriptor: | FRUCTOSE -6-PHOSPHATE, GLYCEROL, SULFATE ION, ... | | Authors: | Swan, M.K, Hansen, T, Schoenheit, P, Davies, C. | | Deposit date: | 2004-08-21 | | Release date: | 2004-12-07 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural basis for phosphomannose isomerase activity in phosphoglucose isomerase from Pyrobaculum aerophilum: a subtle difference between distantly related enzymes.
Biochemistry, 43, 2004
|
|
1CLX
 
 | | CATALYTIC CORE OF XYLANASE A | | Descriptor: | CALCIUM ION, XYLANASE A | | Authors: | Harris, G.W, Jenkins, J.A, Connerton, I, Pickersgill, R.W. | | Deposit date: | 1995-08-31 | | Release date: | 1996-06-20 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Refined crystal structure of the catalytic domain of xylanase A from Pseudomonas fluorescens at 1.8 A resolution.
Acta Crystallogr.,Sect.D, 52, 1996
|
|
3REN
 
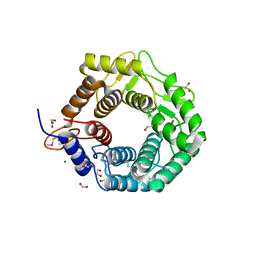 | | CPF_2247, a novel alpha-amylase from Clostridium perfringens | | Descriptor: | 1,2-ETHANEDIOL, Glycosyl hydrolase, family 8, ... | | Authors: | Ficko-Blean, E, Stuart, C.P, Boraston, A.B. | | Deposit date: | 2011-04-04 | | Release date: | 2011-05-18 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural analysis of CPF_2247, a novel alpha-amylase from Clostridium perfringens.
Proteins, 79, 2011
|
|
1NY0
 
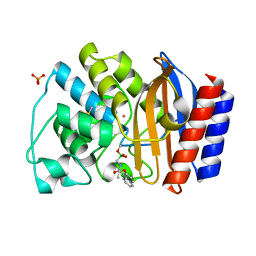 | | Crystal Structure of the complex between M182T mutant of TEM-1 and a boronic acid inhibitor (NBF) | | Descriptor: | Beta-lactamase TEM, PHOSPHATE ION, POTASSIUM ION, ... | | Authors: | Wang, X, Minasov, G, Blazquez, J, Caselli, E, Prati, F, Shoichet, B.K. | | Deposit date: | 2003-02-11 | | Release date: | 2003-08-26 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Recognition and Resistance in TEM beta-lactamase
Biochemistry, 42, 2003
|
|
3CNH
 
 | |
4DCE
 
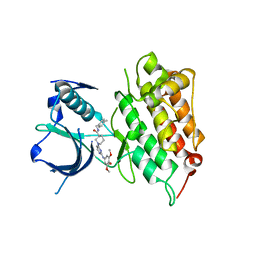 | | Crystal structure of human anaplastic lymphoma kinase in complex with a piperidine-carboxamide inhibitor | | Descriptor: | (3S)-N-(4-methylbenzyl)-1-{2-[(3,4,5-trimethoxyphenyl)amino]pyrimidin-4-yl}piperidine-3-carboxamide, ALK tyrosine kinase receptor | | Authors: | Whittington, D.A, Epstein, L.F, Chen, H. | | Deposit date: | 2012-01-17 | | Release date: | 2012-02-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Rapid development of piperidine carboxamides as potent and selective anaplastic lymphoma kinase inhibitors.
J.Med.Chem., 55, 2012
|
|
5QGT
 
 | | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands) -- Crystal Structure of NUDT7 in complex with FMOPL000609a | | Descriptor: | 1-(2-ethoxyphenyl)piperazine, ACETATE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Krojer, T, Talon, R, Fairhead, M, Diaz Saez, L, Bradley, A.R, Aimon, A, Collins, P, Brandao-Neto, J, Douangamath, A, Ruda, G.F, Szommer, T, Srikannathasan, V, Elkins, J, Spencer, J, London, N, Nelson, A, Brennan, P.E, Huber, K, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F. | | Deposit date: | 2018-05-15 | | Release date: | 2019-03-27 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands)
To Be Published
|
|
3GBA
 
 | | X-ray structure of iGluR5 ligand-binding core (S1S2) in complex with dysiherbaine at 1.35A resolution | | Descriptor: | (2R,3aR,6S,7R,7aR)-2-[(2S)-2-amino-2-carboxyethyl]-6-hydroxy-7-(methylamino)hexahydro-2H-furo[3,2-b]pyran-2-carboxylic acid, CHLORIDE ION, GLYCEROL, ... | | Authors: | Frydenvang, K, Naur, P, Gajhede, M, Kastrup, J.S. | | Deposit date: | 2009-02-19 | | Release date: | 2009-03-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Full Domain Closure of the Ligand-binding Core of the Ionotropic Glutamate Receptor iGluR5 Induced by the High Affinity Agonist Dysiherbaine and the Functional Antagonist 8,9-Dideoxyneodysiherbaine
J.Biol.Chem., 284, 2009
|
|
5A6L
 
 | | High resolution structure of the thermostable glucuronoxylan endo-Beta-1, 4-xylanase, CtXyn30A, from Clostridium thermocellum with two xylobiose units bound | | Descriptor: | CARBOHYDRATE BINDING FAMILY 6, DI(HYDROXYETHYL)ETHER, PHOSPHATE ION, ... | | Authors: | Freire, F, Verma, A.K, Bule, P, Goyal, A, Fontes, C.M.G.A, Najmudin, S. | | Deposit date: | 2015-06-30 | | Release date: | 2016-10-19 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Conservation in the Mechanism of Glucuronoxylan Hydrolysis Revealed by the Structure of Glucuronoxylan Xylano-Hydrolase (Ctxyn30A) from Clostridium Thermocellum
Acta Crystallogr.,Sect.D, 72, 2016
|
|
3RIH
 
 | |
4P10
 
 | | pro-carboxypeptidase U In Complex With 5-(3-aminopropyl)-1-propyl-6,7-dihydro-4H-benzimidazole-5-carboxylic acid | | Descriptor: | (5R)-5-(3-aminopropyl)-1-propyl-4,5,6,7-tetrahydro-1H-benzimidazole-5-carboxylic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, Carboxypeptidase B2, ... | | Authors: | Hallberg, K, Sjogren, T. | | Deposit date: | 2014-02-21 | | Release date: | 2014-08-13 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Design and synthesis of conformationally restricted inhibitors of active thrombin activatable fibrinolysis inhibitor (TAFIa).
Bioorg.Med.Chem., 22, 2014
|
|
3ZMG
 
 | | CRYSTAL STRUCTURE OF BACE-1 IN COMPLEX WITH CHEMICAL LIGAND | | Descriptor: | BETA-SECRETASE 1, DIMETHYL SULFOXIDE, N-[3-[(4R)-2-azanylidene-5,5-bis(fluoranyl)-4-methyl-1,3-oxazinan-4-yl]-4-fluoranyl-phenyl]-5-cyano-pyridine-2-carboxamide, ... | | Authors: | Banner, D.W, Kuglstatter, A, Benz, J, Stihle, M. | | Deposit date: | 2013-02-08 | | Release date: | 2013-05-01 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Beta-Secretase (Bace1) Inhibitors with High in Vivo Efficacy Suitable for Clinical Evaluation in Alzheimer'S Disease.
J.Med.Chem., 56, 2013
|
|
4L71
 
 | | Crystal Structure of Frameshift Suppressor tRNA SufA6 Bound to Codon CCC-A on the Ribosome | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Maehigashi, T, Dunkle, J.A, Dunham, C.M. | | Deposit date: | 2013-06-13 | | Release date: | 2014-08-06 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.900001 Å) | | Cite: | Structural insights into +1 frameshifting promoted by expanded or modification-deficient anticodon stem loops.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
1T0A
 
 | | Crystal Structure of 2C-Methyl-D-Erythritol-2,4-cyclodiphosphate Synthase from Shewanella Oneidensis | | Descriptor: | 2C-methyl-D-erythritol 2,4-cyclodiphosphate synthase, COBALT (II) ION, FARNESYL DIPHOSPHATE, ... | | Authors: | Ni, S, Robinson, H, Marsing, G.C, Bussiere, D.E, Kennedy, M.A. | | Deposit date: | 2004-04-08 | | Release date: | 2004-10-26 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of 2C-methyl-D-erythritol-2,4-cyclodiphosphate synthase from Shewanella oneidensis at 1.6 A: identification of farnesyl pyrophosphate trapped in a hydrophobic cavity.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
7DB5
 
 | | Crystal structure of alpha-L-fucosidase from Vibrio sp. strain EJY3 | | Descriptor: | Alpha-L-fucosidase, GLYCEROL, PHOSPHATE ION | | Authors: | Hong, H, Kim, K.-J. | | Deposit date: | 2020-10-19 | | Release date: | 2021-03-24 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Dual alpha-1,4- and beta-1,4-Glycosidase Activities by the Novel Carbohydrate-Binding Module in alpha-l-Fucosidase from Vibrio sp. Strain EJY3.
J.Agric.Food Chem., 69, 2021
|
|
5QOJ
 
 | | PanDDA analysis group deposition -- Crystal Structure of DCP2 (NUDT20) in complex with FMOCR000171b | | Descriptor: | 1,2-ETHANEDIOL, 2-(thiophen-2-yl)-1H-imidazole, ACETATE ION, ... | | Authors: | Nelson, E.R, Velupillai, S, Talon, R, Collins, P.M, Krojer, T, Wang, D, Brandao-Neto, J, Douangamath, A, Burgess-Brown, N, Arrowsmith, C.H, Bountra, C, Huber, K, von Delft, F. | | Deposit date: | 2019-02-22 | | Release date: | 2019-05-08 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
3G5R
 
 | | Crystal structure of Thermus thermophilus TrmFO in complex with tetrahydrofolate | | Descriptor: | (6S)-5,6,7,8-TETRAHYDROFOLATE, 1,2-ETHANEDIOL, CALCIUM ION, ... | | Authors: | Nishimasu, H, Ishitani, R, Hori, H, Nureki, O. | | Deposit date: | 2009-02-05 | | Release date: | 2009-05-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Atomic structure of a folate/FAD-dependent tRNA T54 methyltransferase
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3D2K
 
 | | Crystal structure of mouse Aurora A (Asn186->Gly, Lys240->Arg, Met302->Leu) in complex with [7-(2-{2-[3-(3-chloro-phenyl)-ureido]-thiazol-5-yl}-ethylamino)-pyrazolo[4,3-d]pyrimidin-1-yl]-acetic acid | | Descriptor: | (7-{[2-(2-{[(3-chlorophenyl)carbamoyl]amino}-1,3-thiazol-5-yl)ethyl]amino}-1H-pyrazolo[4,3-d]pyrimidin-1-yl)acetic acid, serine/threonine kinase 6 | | Authors: | Elling, R.A, Oslob, J.D, Yu, C, Romanowski, M.J. | | Deposit date: | 2008-05-08 | | Release date: | 2009-05-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Discovery of Aurora-A-selective inhibitors
To be Published
|
|
6KPQ
 
 | | Crystal structure of Zika NS2B-NS3 protease with compound 8 | | Descriptor: | 1-[(10~{R},17~{S},20~{S})-17,20-bis(4-azanylbutyl)-4,9,16,19,22-pentakis(oxidanylidene)-3,8,15,18,21-pentazabicyclo[22.3.1]octacosa-1(27),24(28),25-trien-10-yl]guanidine, NS3 protease, Serine protease subunit NS2B | | Authors: | Quek, J.P. | | Deposit date: | 2019-08-15 | | Release date: | 2020-06-17 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Structure-Based Macrocyclization of Substrate Analogue NS2B-NS3 Protease Inhibitors of Zika, West Nile and Dengue viruses.
Chemmedchem, 15, 2020
|
|
3ZX2
 
 | | NTPDase1 in complex with Decavanadate | | Descriptor: | ACETIC ACID, CHLORIDE ION, DECAVANADATE, ... | | Authors: | Zebisch, M, Schaefer, P, Straeter, N. | | Deposit date: | 2011-08-04 | | Release date: | 2011-11-30 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Crystallographic Evidence for a Domain Motion in Rat Nucleoside Triphosphate Diphosphohydrolase (Ntpdase) 1.
J.Mol.Biol., 415, 2012
|
|
3R21
 
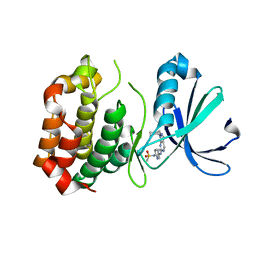 | | Design, synthesis, and biological evaluation of pyrazolopyridine-sulfonamides as potent multiple-mitotic kinase (MMK) inhibitors (Part I) | | Descriptor: | MAGNESIUM ION, N-(2-aminoethyl)-N-{5-[(1-cycloheptyl-1H-pyrazolo[3,4-d]pyrimidin-6-yl)amino]pyridin-2-yl}methanesulfonamide, Serine/threonine-protein kinase 6 | | Authors: | Zhang, L, Fan, J, Chong, J.-H, Cesena, A, Tam, B, Gilson, C, Boykin, C, Wang, D, Marcotte, D, Le Brazidec, J.-Y, Aivazian, D, Piao, J, Lundgren, K, Hong, K, Vu, K, Nguyen, K. | | Deposit date: | 2011-03-11 | | Release date: | 2011-08-10 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Design, synthesis, and biological evaluation of pyrazolopyrimidine-sulfonamides as potent multiple-mitotic kinase (MMK) inhibitors (part I).
Bioorg.Med.Chem.Lett., 21, 2011
|
|
2RHQ
 
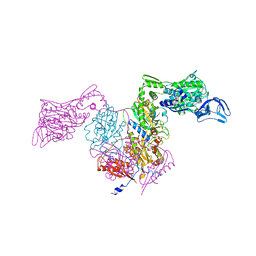 | | PheRS from Staphylococcus haemolyticus- rational protein engineering and inhibitor studies | | Descriptor: | 1-{3-[(4-pyridin-2-ylpiperazin-1-yl)sulfonyl]phenyl}-3-(1,3-thiazol-2-yl)urea, Phenylalanyl-tRNA synthetase alpha chain, Phenylalanyl-tRNA synthetase beta chain, ... | | Authors: | Evdokimov, A.G, Mekel, M. | | Deposit date: | 2007-10-09 | | Release date: | 2007-11-06 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Rational protein engineering in action: the first crystal structure of a phenylalanine tRNA synthetase from Staphylococcus haemolyticus.
J.Struct.Biol., 162, 2008
|
|
2DA9
 
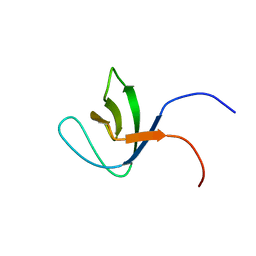 | | Solution structure of the third SH3 domain of SH3-domain kinase binding protein 1 (Regulator of ubiquitous kinase, Ruk) | | Descriptor: | SH3-domain kinase binding protein 1 | | Authors: | Ohnishi, S, Kigawa, T, Saito, K, Koshiba, S, Inoue, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-12-13 | | Release date: | 2006-06-13 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the third SH3 domain of SH3-domain kinase binding protein 1 (Regulator of ubiquitous kinase, Ruk)
To be Published
|
|
6KRG
 
 | | Crystal structure of sfGFP Y182TMSiPhe | | Descriptor: | CHLORIDE ION, GLYCEROL, Green fluorescent protein, ... | | Authors: | Sun, J.P, Wang, J.Y, Zhu, Z.L, He, Q.T, Xiao, P. | | Deposit date: | 2019-08-21 | | Release date: | 2020-09-09 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | DeSiphering receptor core-induced and ligand-dependent conformational changes in arrestin via genetic encoded trimethylsilyl 1 H-NMR probe.
Nat Commun, 11, 2020
|
|
7FIY
 
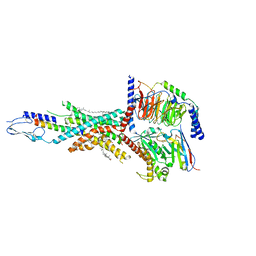 | | Cryo-EM structure of the tirzepatide-bound human GIPR-Gs complex | | Descriptor: | CHOLESTEROL, Gastric inhibitory polypeptide receptor,Gastric inhibitory polypeptide receptor,human glucose-dependent insulinotropic polypeptide receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Zhao, F.H, Zhou, Q.T, Cong, Z.T, Hang, K.N, Zou, X.Y, Zhang, C, Chen, Y, Dai, A.T, Liang, A.Y, Ming, Q.Q, Wang, M, Chen, L.N, Xu, P.Y, Chang, R.L, Feng, W.B, Xia, T, Zhang, Y, Wu, B.L, Yang, D.H, Zhao, L.H, Xu, H.E, Wang, M.W. | | Deposit date: | 2021-08-01 | | Release date: | 2022-03-02 | | Last modified: | 2022-03-16 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural insights into multiplexed pharmacological actions of tirzepatide and peptide 20 at the GIP, GLP-1 or glucagon receptors.
Nat Commun, 13, 2022
|
|
