7NHZ
 
 | |
7NPL
 
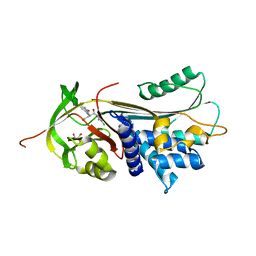 | | ALPHA-1 ANTITRYPSIN (C232S) COMPLEXED WITH cmpd 11 | | Descriptor: | Alpha-1-antitrypsin, GLYCEROL, N-((1S,2R)-1-(3-chloro-2-methylphenyl)-1-hydroxypentan-2-yl)-2-oxoindoline-4-carboxamide | | Authors: | Chung, C. | | Deposit date: | 2021-02-27 | | Release date: | 2021-04-07 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | The development of highly potent and selective small molecule correctors of Z alpha 1 -antitrypsin misfolding.
Bioorg.Med.Chem.Lett., 41, 2021
|
|
7NG3
 
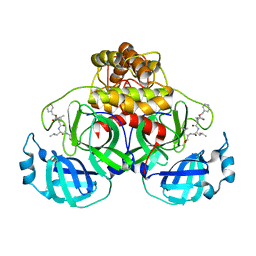 | | Crystal structure of MG-132 covalently bound to the main protease (3CLpro/Mpro) of SARS-CoV-2 in spacegroup P1. | | Descriptor: | 3C-like proteinase, CHLORIDE ION, N-[(benzyloxy)carbonyl]-L-leucyl-N-[(2S)-1-hydroxy-4-methylpentan-2-yl]-L-leucinamide | | Authors: | Costanzi, E, Demitri, N, Giabbai, B, Storici, P. | | Deposit date: | 2021-02-08 | | Release date: | 2021-04-07 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and Biochemical Analysis of the Dual Inhibition of MG-132 against SARS-CoV-2 Main Protease (Mpro/3CLpro) and Human Cathepsin-L.
Int J Mol Sci, 22, 2021
|
|
7NGC
 
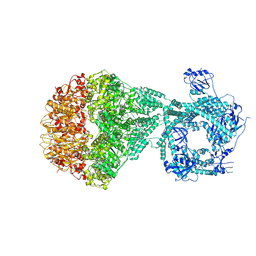 | | P2a-state of wild type human mitochondrial LONP1 protease with bound substrate protein and in presence of ATPgS | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Lon protease homolog, mitochondrial, ... | | Authors: | Mohammed, I, Schmitz, K.A, Schenck, N, Maier, T, Abrahams, J.P. | | Deposit date: | 2021-02-09 | | Release date: | 2021-04-07 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (7.5 Å) | | Cite: | Catalytic cycling of human mitochondrial Lon protease.
Structure, 30, 2022
|
|
7NN6
 
 | |
7NG6
 
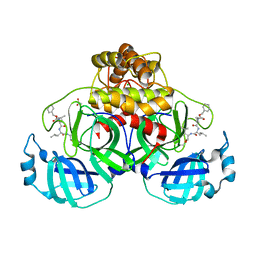 | | Crystal structure of MG-132 covalently bound to the main protease (3CLpro/Mpro) of SARS-CoV-2 in spacegroup P1 in absence of DTT. | | Descriptor: | 3C-like proteinase, ACETATE ION, CHLORIDE ION, ... | | Authors: | Costanzi, E, Demitri, N, Giabbai, B, Storici, P. | | Deposit date: | 2021-02-08 | | Release date: | 2021-04-07 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Structural and Biochemical Analysis of the Dual Inhibition of MG-132 against SARS-CoV-2 Main Protease (Mpro/3CLpro) and Human Cathepsin-L.
Int J Mol Sci, 22, 2021
|
|
3CZH
 
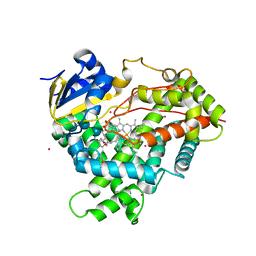 | | Crystal structure of CYP2R1 in complex with vitamin D2 | | Descriptor: | (3S,5Z,7E,22E)-9,10-secoergosta-5,7,10,22-tetraen-3-ol, Cycloheptakis-(1-4)-(alpha-D-glucopyranose), Cytochrome P450 2R1, ... | | Authors: | Strushkevich, N.V, Tempel, W, Gilep, A.A, Loppnau, P, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Wilkstrom, M, Bochkarev, A, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-04-29 | | Release date: | 2008-08-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of CYP2R1 in complex with vitamin D2.
To be Published
|
|
7NPK
 
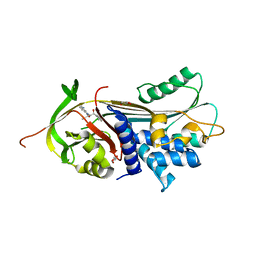 | | ALPHA-1 ANTITRYPSIN C232S COMPLEXED WITH CMPD3 | | Descriptor: | Alpha-1-antitrypsin, GLYCEROL, N-((1S,2R)-1-hydroxy-1-(o-tolyl)pentan-2-yl)-2-oxo-2,3-dihydrobenzo[d]oxazole-5-carboxamide | | Authors: | Chung, C. | | Deposit date: | 2021-02-27 | | Release date: | 2021-04-07 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | The development of highly potent and selective small molecule correctors of Z alpha 1 -antitrypsin misfolding.
Bioorg.Med.Chem.Lett., 41, 2021
|
|
7NNG
 
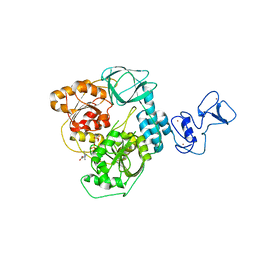 | | Crystal structure of the SARS-CoV-2 helicase in complex with Z2327226104 | | Descriptor: | 1-(2-methylphenyl)-1,2,3-triazole-4-carboxylic acid, PHOSPHATE ION, SARS-CoV-2 helicase NSP13, ... | | Authors: | Newman, J.A, Yosaatmadja, Y, Douangamath, A, Bountra, C, Gileadi, O. | | Deposit date: | 2021-02-24 | | Release date: | 2021-04-07 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Structure, mechanism and crystallographic fragment screening of the SARS-CoV-2 NSP13 helicase.
Nat Commun, 12, 2021
|
|
7NQJ
 
 | | C-TERMINAL BROMODOMAIN OF HUMAN BRD2 WITH N-ethyl-2-(1-methyl-1H-1,2,3-triazol-4-yl)-6-(1-phenylethyl)isonicotinamide | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 2, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Chung, C. | | Deposit date: | 2021-03-01 | | Release date: | 2021-04-07 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.734 Å) | | Cite: | Template-Hopping Approach Leads to Potent, Selective, and Highly Soluble Bromo and Extraterminal Domain (BET) Second Bromodomain (BD2) Inhibitors.
J.Med.Chem., 64, 2021
|
|
7NRN
 
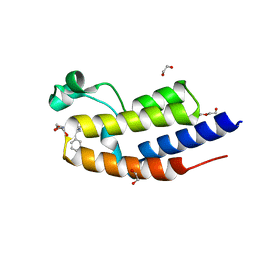
 | | NMR structure of GIPC1-GH2 domain | | Descriptor: | PDZ domain-containing protein GIPC1 | | Authors: | Barthe, P, Roumestand, C. | | Deposit date: | 2021-03-04 | | Release date: | 2021-04-14 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Pressure and Chemical Unfolding of an alpha-Helical Bundle Protein: The GH2 Domain of the Protein Adaptor GIPC1.
Int J Mol Sci, 22, 2021
|
|
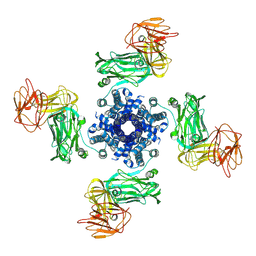
7NTX
 
 | |
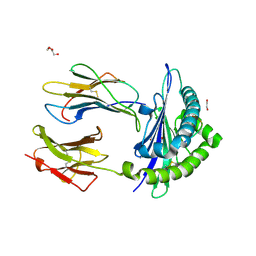
7NUI
 
 | |
7NRD
 
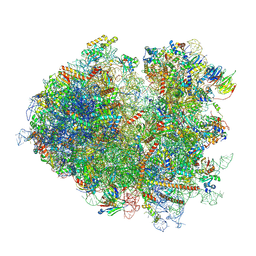 | | Structure of the yeast Gcn1 bound to a colliding stalled 80S ribosome with MBF1, A/P-tRNA and P/E-tRNA | | Descriptor: | 25S rRNA (3184-MER), 40S ribosomal protein S0-A, 40S ribosomal protein S1-A, ... | | Authors: | Pochopien, A.A, Beckert, B, Wilson, D.N. | | Deposit date: | 2021-03-03 | | Release date: | 2021-04-14 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (4.36 Å) | | Cite: | Structure of Gcn1 bound to stalled and colliding 80S ribosomes.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7NQ6
 
 | |
7NXZ
 
 | |
7O1Q
 
 | | Amyloid beta oligomer displayed on the alpha hemolysin scaffold | | Descriptor: | Alpha-hemolysin hybridized Abeta | | Authors: | Wu, J, Blum, T.B, Farrell, D.P, DiMaio, F, Abrahams, J.P, Luo, J. | | Deposit date: | 2021-03-30 | | Release date: | 2021-04-14 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryo-electron Microscopy Imaging of Alzheimer's Amyloid-beta 42 Oligomer Displayed on a Functionally and Structurally Relevant Scaffold.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
7O39
 
 | |
7NO8
 
 | | Structure of the mature RSV CA lattice: Group I, pentamer-hexamer interface, class 1"6 | | Descriptor: | Capsid protein p27, alternate cleaved 1 | | Authors: | Obr, M, Ricana, C.L, Nikulin, N, Feathers, J.-P.R, Klanschnig, M, Thader, A, Johnson, M.C, Vogt, V.M, Schur, F.K.M, Dick, R.A. | | Deposit date: | 2021-02-25 | | Release date: | 2021-04-21 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (7.9 Å) | | Cite: | Structure of the mature Rous sarcoma virus lattice reveals a role for IP6 in the formation of the capsid hexamer.
Nat Commun, 12, 2021
|
|
7NOM
 
 | | Structure of the mature RSV CA lattice: Group IV, hexamer-hexamer interface, class 4'Beta | | Descriptor: | Capsid protein p27, alternate cleaved 1 | | Authors: | Obr, M, Ricana, C.L, Nikulin, N, Feathers, J.-P.R, Klanschnig, M, Thader, A, Johnson, M.C, Vogt, V.M, Schur, F.K.M, Dick, R.A. | | Deposit date: | 2021-02-25 | | Release date: | 2021-04-21 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (6.7 Å) | | Cite: | Structure of the mature Rous sarcoma virus lattice reveals a role for IP6 in the formation of the capsid hexamer.
Nat Commun, 12, 2021
|
|
7NOG
 
 | | Structure of the mature RSV CA lattice: Group III, hexamer-hexamer interface, class 4'5 | | Descriptor: | Capsid protein p27, alternate cleaved 1 | | Authors: | Obr, M, Ricana, C.L, Nikulin, N, Feathers, J.-P.R, Klanschnig, M, Thader, A, Johnson, M.C, Vogt, V.M, Schur, F.K.M, Dick, R.A. | | Deposit date: | 2021-02-25 | | Release date: | 2021-04-21 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (7.8 Å) | | Cite: | Structure of the mature Rous sarcoma virus lattice reveals a role for IP6 in the formation of the capsid hexamer.
Nat Commun, 12, 2021
|
|
7NO0
 
 | | Structure of the mature RSV CA lattice: T=1 CA icosahedron | | Descriptor: | Capsid protein p27, alternate cleaved 1 | | Authors: | Obr, M, Ricana, C.L, Nikulin, N, Feathers, J.-P.R, Klanschnig, M, Thader, A, Johnson, M.C, Vogt, V.M, Schur, F.K.M, Dick, R.A. | | Deposit date: | 2021-02-25 | | Release date: | 2021-04-21 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structure of the mature Rous sarcoma virus lattice reveals a role for IP6 in the formation of the capsid hexamer.
Nat Commun, 12, 2021
|
|
7NOB
 
 | | Structure of the mature RSV CA lattice: Group II, hexamer-hexamer interface, class 2'6 | | Descriptor: | Capsid protein p27, alternate cleaved 1 | | Authors: | Obr, M, Ricana, C.L, Nikulin, N, Feathers, J.-P.R, Klanschnig, M, Thader, A, Johnson, M.C, Vogt, V.M, Schur, F.K.M, Dick, R.A. | | Deposit date: | 2021-02-25 | | Release date: | 2021-04-21 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (6.7 Å) | | Cite: | Structure of the mature Rous sarcoma virus lattice reveals a role for IP6 in the formation of the capsid hexamer.
Nat Commun, 12, 2021
|
|
7NO2
 
 | | Structure of the mature RSV CA lattice: hexamer derived from tubes (C2-symmetric) | | Descriptor: | Capsid protein p27, alternate cleaved 1 | | Authors: | Obr, M, Ricana, C.L, Nikulin, N, Feathers, J.-P.R, Klanschnig, M, Thader, A, Johnson, M.C, Vogt, V.M, Schur, F.K.M, Dick, R.A. | | Deposit date: | 2021-02-25 | | Release date: | 2021-04-21 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Structure of the mature Rous sarcoma virus lattice reveals a role for IP6 in the formation of the capsid hexamer.
Nat Commun, 12, 2021
|
|
7NOI
 
 | | Structure of the mature RSV CA lattice: Group IV, hexamer-hexamer interface, class 3'Alpha | | Descriptor: | Capsid protein p27, alternate cleaved 1 | | Authors: | Obr, M, Ricana, C.L, Nikulin, N, Feathers, J.-P.R, Klanschnig, M, Thader, A, Johnson, M.C, Vogt, V.M, Schur, F.K.M, Dick, R.A. | | Deposit date: | 2021-02-25 | | Release date: | 2021-04-21 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (7.2 Å) | | Cite: | Structure of the mature Rous sarcoma virus lattice reveals a role for IP6 in the formation of the capsid hexamer.
Nat Commun, 12, 2021
|
|
