1XBD
 
 | | INTERNAL XYLAN BINDING DOMAIN FROM CELLULOMONAS FIMI XYLANASE D, NMR, 5 STRUCTURES | | Descriptor: | XYLANASE D | | Authors: | Simpson, P.J, Bolam, D.N, Cooper, A, Ciruela, A, Hazlewood, G.P, Gilbert, H.J, Williamson, M.P. | | Deposit date: | 1998-10-16 | | Release date: | 1999-07-21 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | A family IIb xylan-binding domain has a similar secondary structure to a homologous family IIa cellulose-binding domain but different ligand specificity.
Structure Fold.Des., 7, 1999
|
|
1XBF
 
 | | X-RAY STRUCTURE NORTHEAST STRUCTURAL GENOMICS CONSORTIUM TARGET CAR10 FROM C. ACETOBUTYLICUM | | Descriptor: | Clostridium acetobutylicum Q97KL0, SULFATE ION | | Authors: | Kuzin, A.P, Chen, Y, Vorobiev, S, Yong, W, Acton, T, Ho, C.-K, Conover, K, Cooper, B, Ciano, M, Xiao, R, Montelione, G, Tong, L, Hunt, J, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2004-08-30 | | Release date: | 2004-09-14 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-RAY STRUCTURE NORTHEAST STRUCTURAL GENOMICS CONSORTIUM TARGET CAR10 FROM C. ACETOBUTYLICUM
To be published
|
|
1XBH
 
 | | A BETA-HAIRPIN MIMIC FROM FCERI-ALPHA-CYCLO(L-262) | | Descriptor: | PROTEIN (CYCLO(L-262)) | | Authors: | Mcdonnell, J.M, Fushman, D, Cahill, S.M, Sutton, B.J, Cowburn, D. | | Deposit date: | 1999-02-17 | | Release date: | 1999-02-21 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution Structures of FceRI Alpha-Chain Mimics: A Beta-Hairpin Peptide and Its Retroenantiomer
J.Am.Chem.Soc., 119, 1997
|
|
1XBI
 
 | | High resolution structure of Methanocaldococcus jannaschii L7AE | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, 50S ribosomal protein L7Ae | | Authors: | Brown II, B.A, Suryadi, J, Lieberman, D.V, Tran, E.J, Maxwell, E.S. | | Deposit date: | 2004-08-30 | | Release date: | 2005-08-09 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | The Crystal Structure of the Methanocaldococcus jannaschii Multifunctional L7Ae RNA-Binding Protein Reveals an Induced-Fit Interaction with the Box C/D RNAs.
Biochemistry, 44, 2005
|
|
1XBL
 
 | | NMR STRUCTURE OF THE J-DOMAIN (RESIDUES 2-76) IN THE ESCHERICHIA COLI N-TERMINAL FRAGMENT (RESIDUES 2-108) OF THE MOLECULAR CHAPERONE DNAJ, 20 STRUCTURES | | Descriptor: | DNAJ | | Authors: | Pellecchia, M, Szyperski, T, Wall, D, Georgopoulos, C, Wuthrich, K. | | Deposit date: | 1996-10-07 | | Release date: | 1997-01-11 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the J-domain and the Gly/Phe-rich region of the Escherichia coli DnaJ chaperone.
J.Mol.Biol., 260, 1996
|
|
1XBN
 
 | |
1XBO
 
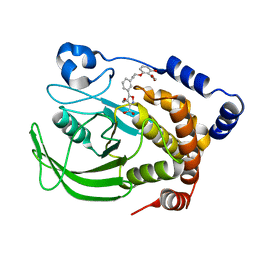 | | PTP1B complexed with Isoxazole Carboxylic Acid | | Descriptor: | 5-(3-{3-[3-HYDROXY-2-(METHOXYCARBONYL)PHENOXY]PROPENYL}PHENYL)-4-(HYDROXYMETHYL)ISOXAZOLE-3-CARBOXYLIC ACID, Protein-tyrosine phosphatase, non-receptor type 1 | | Authors: | Zhao, H, Liu, G, Xin, Z, Serby, M, Pei, Z, Szczepankiewicz, B.G, Hajduk, P.J, Abad-Zapatero, C, Hutchins, C.W, Lubben, T.H, Ballaron, S.J, Hassach, D.L, Kaszubska, W, Rondinone, C.M, Trevillyan, J.M, Jirousek, M.R. | | Deposit date: | 2004-08-31 | | Release date: | 2004-10-19 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Isoxazole carboxylic acids as protein tyrosine phosphatase 1B (PTP1B) inhibitors.
Bioorg.Med.Chem.Lett., 14, 2004
|
|
1XBP
 
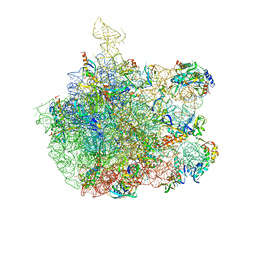 | | Inhibition of peptide bond formation by pleuromutilins: The structure of the 50S ribosomal subunit from Deinococcus radiodurans in complex with Tiamulin | | Descriptor: | 23S RIBOSOMAL RNA, 50S ribosomal protein L11, 50S ribosomal protein L13, ... | | Authors: | Schluenzen, F, Pyetan, E, Fucini, P, Yonath, A, Harms, J.M. | | Deposit date: | 2004-08-31 | | Release date: | 2005-03-01 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Inhibition of peptide bond formation by pleuromutilins: the structure of the 50S ribosomal subunit from Deinococcus radiodurans in complex with tiamulin.
Mol.Microbiol., 54, 2004
|
|
1XBR
 
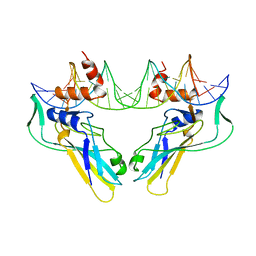 | | T DOMAIN FROM XENOPUS LAEVIS BOUND TO DNA | | Descriptor: | DNA (5'-D(*AP*AP*TP*TP*TP*CP*AP*CP*AP*CP*CP*TP*AP*GP*GP*TP*G P*TP*GP*AP*AP*AP* TP*T)-3'), PROTEIN (T PROTEIN) | | Authors: | Muller, C.W. | | Deposit date: | 1997-07-16 | | Release date: | 1998-01-16 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystallographic structure of the T domain-DNA complex of the Brachyury transcription factor.
Nature, 389, 1997
|
|
1XBS
 
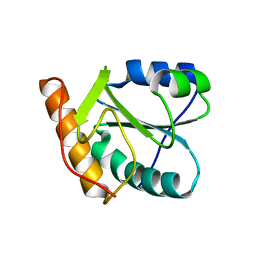 | | Crystal structure of human dim2: a dim1-like protein | | Descriptor: | Dim1-like protein | | Authors: | Simeoni, F, Arvai, A, Hopfner, K.-P, Bello, P, Gondeau, C, Heitz, F, Tainer, J, Divita, G. | | Deposit date: | 2004-08-31 | | Release date: | 2005-08-30 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Biochemical Characterization and Crystal Structure of a Dim1 Family Associated Protein: Dim2
Biochemistry, 44, 2005
|
|
1XBT
 
 | | Crystal Structure of Human Thymidine Kinase 1 | | Descriptor: | MAGNESIUM ION, THYMIDINE-5'-TRIPHOSPHATE, Thymidine kinase, ... | | Authors: | Welin, M, Kosinska, U, Mikkelsen, N.E, Carnrot, C, Zhu, C, Wang, L, Eriksson, S, Munch-Petersen, B, Eklund, H. | | Deposit date: | 2004-08-31 | | Release date: | 2004-12-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structures of thymidine kinase 1 of human and mycoplasmic origin
Proc.Natl.Acad.Sci.Usa, 101, 2004
|
|
1XBU
 
 | | Streptomyces griseus aminopeptidase complexed with p-iodo-D-phenylalanine | | Descriptor: | Aminopeptidase, CALCIUM ION, P-IODO-D-PHENYLALANINE, ... | | Authors: | Reiland, V, Gilboa, R, Spungin-Bialik, A, Schomburg, D, Shoham, Y, Blumberg, S, Shoham, G. | | Deposit date: | 2004-08-31 | | Release date: | 2005-10-04 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Streptomyces griseus aminopeptidase complexed with p-iodo-D-phenylalanine
To be Published
|
|
1XBV
 
 | | Crystal structure of 3-keto-L-gulonate 6-phosphate decarboxylase with bound D-ribulose 5-phosphate | | Descriptor: | 3-keto-L-gulonate 6-phosphate decarboxylase, MAGNESIUM ION, RIBULOSE-5-PHOSPHATE | | Authors: | Wise, E.L, Yew, W.S, Akana, J, Gerlt, J.A, Rayment, I. | | Deposit date: | 2004-08-31 | | Release date: | 2005-04-26 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Evolution of enzymatic activities in the orotidine 5'-monophosphate decarboxylase suprafamily: structural basis for catalytic promiscuity in wild-type and designed mutants of 3-keto-L-gulonate 6-phosphate decarboxylase
Biochemistry, 44, 2005
|
|
1XBW
 
 | | 1.9A Crystal Structure of the protein isdG from Staphylococcus aureus aureus, Structural genomics, MCSG | | Descriptor: | hypothetical protein isdG | | Authors: | Zhang, R, Wu, R, Joachimiak, G, Schneewind, O, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-08-31 | | Release date: | 2004-10-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Staphylococcus aureus IsdG and IsdI, heme-degrading enzymes with structural similarity to monooxygenases.
J.Biol.Chem., 280, 2005
|
|
1XBX
 
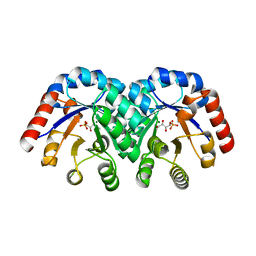 | | Structure of 3-keto-L-gulonate 6-phosphate decarboxylase E112D/R139V/T169A mutant with bound D-ribulose 5-phosphate | | Descriptor: | 3-keto-L-gulonate 6-phosphate decarboxylase, 5-O-phosphono-L-ribulose, MAGNESIUM ION, ... | | Authors: | Wise, E.L, Yew, W.S, Akana, J, Gerlt, J.A, Rayment, I. | | Deposit date: | 2004-08-31 | | Release date: | 2005-04-26 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Evolution of enzymatic activities in the orotidine 5'-monophosphate decarboxylase suprafamily: structural basis for catalytic promiscuity in wild-type and designed mutants of 3-keto-L-gulonate 6-phosphate decarboxylase
Biochemistry, 44, 2005
|
|
1XBY
 
 | | Structure of 3-keto-L-gulonate 6-phosphate decarboxylase E112D/T169A mutant with bound D-ribulose 5-phosphate | | Descriptor: | 3-keto-L-gulonate 6-phosphate decarboxylase, MAGNESIUM ION, RIBULOSE-5-PHOSPHATE | | Authors: | Wise, E.L, Yew, W.S, Akana, J, Gerlt, J.A, Rayment, I. | | Deposit date: | 2004-08-31 | | Release date: | 2005-04-26 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Evolution of enzymatic activities in the orotidine 5'-monophosphate decarboxylase suprafamily: structural basis for catalytic promiscuity in wild-type and designed mutants of 3-keto-L-gulonate 6-phosphate decarboxylase
Biochemistry, 44, 2005
|
|
1XBZ
 
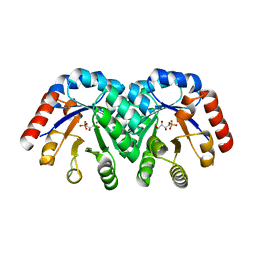 | | Crystal structure of 3-keto-L-gulonate 6-phosphate decarboxylase E112D/R139V/T169A mutant with bound L-xylulose 5-phosphate | | Descriptor: | 3-Keto-L-Gulonate 6-Phosphate Decarboxylase, L-XYLULOSE 5-PHOSPHATE, MAGNESIUM ION | | Authors: | Wise, E.L, Yew, W.S, Akana, J, Gerlt, J.A, Rayment, I. | | Deposit date: | 2004-08-31 | | Release date: | 2005-04-26 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Evolution of enzymatic activities in the orotidine 5'-monophosphate decarboxylase suprafamily: structural basis for catalytic promiscuity in wild-type and designed mutants of 3-keto-L-gulonate 6-phosphate decarboxylase
Biochemistry, 44, 2005
|
|
1XC0
 
 | | Twenty Lowest Energy Structures of Pa4 by Solution NMR | | Descriptor: | Pardaxin P-4 | | Authors: | Porcelli, F, Buck, B, Lee, D.-K, Hallock, K.J, Ramamoorthy, A, Veglia, G. | | Deposit date: | 2004-08-31 | | Release date: | 2004-09-28 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure and orientation of pardaxin determined by NMR experiments in model membranes
J.Biol.Chem., 279, 2004
|
|
1XC1
 
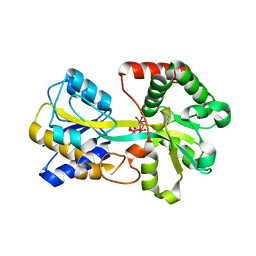 | | Oxo Zirconium(IV) Cluster in the Ferric Binding Protein (FBP) | | Descriptor: | OXO ZIRCONIUM(IV) CLUSTER, periplasmic iron-binding protein | | Authors: | Zhong, W, Alexeev, D, Harvey, I, Guo, M, Hunter, D.J.B, Zhu, H, Campopiano, D.J, Sadler, P.J. | | Deposit date: | 2004-08-31 | | Release date: | 2004-09-21 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Assembly of an Oxo-Zirconium(IV) Cluster in a Protein Cleft
Angew.Chem.Int.Ed.Engl., 43, 2004
|
|
1XC3
 
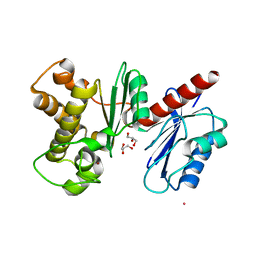 | | Structure of a Putative Fructokinase from Bacillus subtilis | | Descriptor: | GLYCEROL, PLATINUM (II) ION, Putative fructokinase, ... | | Authors: | Cuff, M.E, Quartey, P, Lezondra, L, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-08-31 | | Release date: | 2004-10-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural studies of ROK fructokinase YdhR from Bacillus subtilis: insights into substrate binding and fructose specificity.
J.Mol.Biol., 406, 2011
|
|
1XC4
 
 | |
1XC5
 
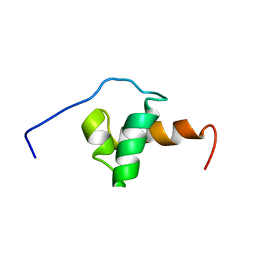 | | Solution Structure of the SMRT Deacetylase Activation Domain | | Descriptor: | Nuclear receptor corepressor 2 | | Authors: | Codina, A, Love, J.D, Li, Y, Lazar, M.A, Neuhaus, D, Schwabe, J.W.R. | | Deposit date: | 2004-09-01 | | Release date: | 2005-05-03 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structural insights into the interaction and activation of histone deacetylase 3 by nuclear receptor corepressors
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
1XC6
 
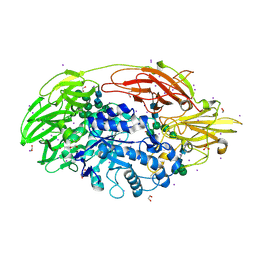 | | Native Structure Of Beta-Galactosidase from Penicillium sp. in complex with Galactose | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Rojas, A.L, Nagem, R.A.P, Neustroev, K.N, Arand, M, Adamska, M, Eneyskaya, E.V, Kulminskaya, A.A, Garratt, R.C, Golubev, A.M, Polikarpov, I. | | Deposit date: | 2004-09-01 | | Release date: | 2004-11-02 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structures of beta-Galactosidase from Penicillium sp. and its Complex with Galactose
J.Mol.Biol., 343, 2004
|
|
1XC7
 
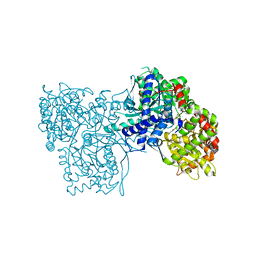 | | Binding of beta-D-glucopyranosyl bismethoxyphosphoramidate to glycogen phosphorylase b: Kinetic and crystallographic studies | | Descriptor: | Glycogen phosphorylase, muscle form, N-(dimethoxyphosphoryl)-beta-D-glucopyranosylamine, ... | | Authors: | Chrysina, E.D, Kosmopoulou, M.N, Kardakaris, R, Bischler, N, Leonidas, D.D, Kannan, T, Loganathan, D, Oikonomakos, N.G. | | Deposit date: | 2004-09-01 | | Release date: | 2005-02-08 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Binding of beta-d-glucopyranosyl bismethoxyphosphoramidate to glycogen phosphorylase b: kinetic and crystallographic studies
Bioorg.Med.Chem., 13, 2005
|
|
1XC8
 
 | | CRYSTAL STRUCTURE COMPLEX BETWEEN THE WILD-TYPE LACTOCOCCUS LACTIS FPG (MUTM) AND A FAPY-DG CONTAINING DNA | | Descriptor: | 5'-D(*CP*TP*CP*TP*TP*TP*(FOX)P*TP*TP*TP*CP*TP*CP*G)-3', 5'-D(*GP*CP*GP*AP*GP*AP*AP*AP*CP*AP*AP*AP*GP*A)-3', Formamidopyrimidine-DNA glycosylase, ... | | Authors: | Coste, F, Ober, M, Carell, T, Boiteux, S, Zelwer, C, Castaing, B. | | Deposit date: | 2004-09-01 | | Release date: | 2005-09-06 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural basis for the recognition of the FapydG lesion (2,6-diamino-4-hydroxy-5-formamidopyrimidine) by formamidopyrimidine-DNA glycosylase.
J.Biol.Chem., 279, 2004
|
|
