2EAV
 
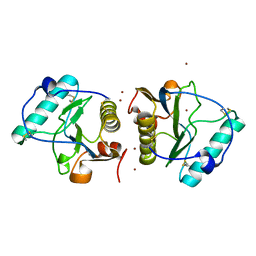 | |
1L27
 
 | |
1LRL
 
 | | Crystal Structure of UDP-Galactose 4-Epimerase Mutant Y299C Complexed with UDP-Glucose | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SODIUM ION, TRIETHYLENE GLYCOL, ... | | Authors: | Thoden, J.B, Henderson, J.M, Fridovich-Keil, J.L, Holden, H.M. | | Deposit date: | 2002-05-15 | | Release date: | 2002-07-26 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural analysis of the Y299C mutant of Escherichia coli UDP-galactose 4-epimerase. Teaching an old dog new tricks.
J.Biol.Chem., 277, 2002
|
|
1L62
 
 | |
1H31
 
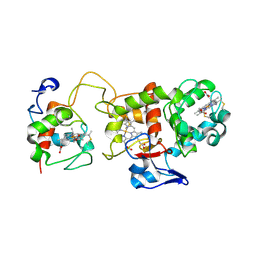 | | Oxidised SoxAX complex from Rhodovulum sulfidophilum | | Descriptor: | CYTOCHROME C, DIHEME CYTOCHROME C, HEME C | | Authors: | Bamford, V.A, Bruno, S, Rasmussen, T, Appia-Ayme, C, Cheesman, M.R, Berks, B.C, Hemmings, A.M. | | Deposit date: | 2002-08-21 | | Release date: | 2002-11-07 | | Last modified: | 2013-03-06 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structural Basis for the Oxidation of Thiosulfate by a Sulfur Cycle Enzyme
Embo J., 21, 2002
|
|
1L73
 
 | |
2F8B
 
 | |
1L11
 
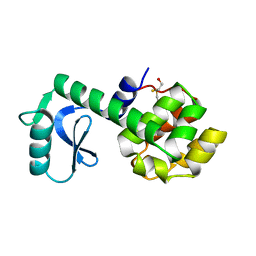 | | CONTRIBUTIONS OF HYDROGEN BONDS OF THR 157 TO THE THERMODYNAMIC STABILITY OF PHAGE T4 LYSOZYME | | Descriptor: | BETA-MERCAPTOETHANOL, T4 LYSOZYME | | Authors: | Dao-Pin, S, Wilson, K, Alber, T, Matthews, B.W. | | Deposit date: | 1988-02-05 | | Release date: | 1988-04-16 | | Last modified: | 2022-11-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Contributions of hydrogen bonds of Thr 157 to the thermodynamic stability of phage T4 lysozyme.
Nature, 330, 1987
|
|
1H1N
 
 | |
2FAD
 
 | | Crystal structure of E. coli heptanoyl-ACP | | Descriptor: | Acyl carrier protein, S-(2-{[N-(2-HYDROXY-4-{[HYDROXY(OXIDO)PHOSPHINO]OXY}-3,3-DIMETHYLBUTANOYL)-BETA-ALANYL]AMINO}ETHYL) HEPTANETHIOATE, SODIUM ION, ... | | Authors: | Roujeinikova, A. | | Deposit date: | 2005-12-07 | | Release date: | 2006-09-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural Studies of Fatty Acyl-(Acyl Carrier Protein) Thioesters Reveal a Hydrophobic Binding Cavity that Can Expand to Fit Longer Substrates.
J.Mol.Biol., 365, 2007
|
|
1L14
 
 | |
1H4P
 
 | | Crystal structure of exo-1,3-beta glucanse from Saccharomyces cerevisiae | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLUCAN 1,3-BETA-GLUCOSIDASE I/II, GLYCEROL, ... | | Authors: | Ferguson, A.D. | | Deposit date: | 2001-05-11 | | Release date: | 2003-10-02 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The Er Protein Folding Sensor Udp-Glucose Glycoprotein:Glucosyltransferase Modifies Substrates Distant to Local Changes in Glycoprotein Conformation.
Nat.Struct.Mol.Biol., 11, 2004
|
|
1L1D
 
 | | Crystal structure of the C-terminal methionine sulfoxide reductase domain (MsrB) of N. gonorrhoeae pilB | | Descriptor: | CACODYLATE ION, peptide methionine sulfoxide reductase | | Authors: | Lowther, W.T, Weissbach, H, Etienne, F, Brot, N, Matthews, B.W. | | Deposit date: | 2002-02-15 | | Release date: | 2002-05-01 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The mirrored methionine sulfoxide reductases of Neisseria gonorrhoeae pilB.
Nat.Struct.Biol., 9, 2002
|
|
1L57
 
 | |
3BS1
 
 | | Structure of the Staphylococcus aureus AgrA LytTR Domain Bound to DNA Reveals a Beta Fold with a Novel Mode of Binding | | Descriptor: | Accessory gene regulator protein A, DNA (5'-D(*DAP*DAP*(BRU)P*DAP*DCP*DTP*DTP*DAP*DAP*DCP*DTP*DGP*DTP*DTP*DAP*DA)-3'), DNA (5'-D(*DTP*DTP*DTP*DAP*DAP*DCP*DAP*DGP*DTP*DTP*DAP*DAP*DGP*(BRU)P*DAP*DT)-3'), ... | | Authors: | Sidote, D.J, Barbieri, C, Wu, T, Stock, A.M. | | Deposit date: | 2007-12-21 | | Release date: | 2008-04-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of the Staphylococcus aureus AgrA LytTR Domain Bound to DNA Reveals a Beta Fold with an Unusual Mode of Binding.
Structure, 16, 2008
|
|
1L46
 
 | |
1HDR
 
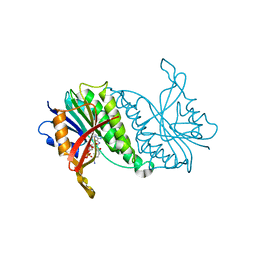 | | THE CRYSTALLOGRAPHIC STRUCTURE OF A HUMAN DIHYDROPTERIDINE REDUCTASE NADH BINARY COMPLEX EXPRESSED IN ESCHERICHIA COLI BY A CDNA CONSTRUCTED FROM ITS RAT HOMOLOGUE | | Descriptor: | DIHYDROPTERIDINE REDUCTASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Varughese, K.I, Su, Y, Xuong, N.H, Whiteley, J.M. | | Deposit date: | 1993-08-18 | | Release date: | 1994-08-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The crystallographic structure of a human dihydropteridine reductase NADH binary complex expressed in Escherichia coli by a cDNA constructed from its rat homologue.
J.Biol.Chem., 268, 1993
|
|
1HEO
 
 | |
1L81
 
 | |
1L65
 
 | |
1HEW
 
 | |
1L88
 
 | |
1L72
 
 | |
1L86
 
 | |
4O5B
 
 | |
