1C0M
 
 | |
1C0N
 
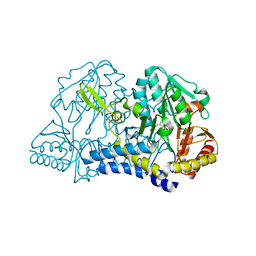 | | CSDB PROTEIN, NIFS HOMOLOGUE | | Descriptor: | ACETIC ACID, PROTEIN (CSDB PROTEIN), PYRIDOXAL-5'-PHOSPHATE | | Authors: | Fujii, T, Maeda, M, Mihara, H, Kurihara, T, Esaki, N, Hata, Y. | | Deposit date: | 1999-07-17 | | Release date: | 2000-07-17 | | Last modified: | 2017-10-04 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of a NifS homologue: X-ray structure analysis of CsdB, an Escherichia coli counterpart of mammalian selenocysteine lyase
Biochemistry, 39, 2000
|
|
1C0O
 
 | |
1C0P
 
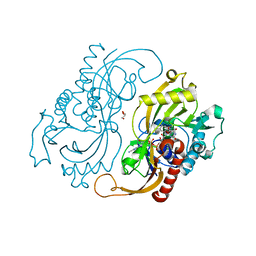 | | D-AMINO ACIC OXIDASE IN COMPLEX WITH D-ALANINE AND A PARTIALLY OCCUPIED BIATOMIC SPECIES | | Descriptor: | D-ALANINE, D-AMINO ACID OXIDASE, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Umhau, S, Pollegioni, L, Molla, G, Diederichs, K, Welte, W, Pilone, S.M, Ghisla, S. | | Deposit date: | 1999-07-19 | | Release date: | 2000-11-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | The x-ray structure of D-amino acid oxidase at very high resolution identifies the chemical mechanism of flavin-dependent substrate dehydrogenation.
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
1C0Q
 
 | | COMPLEX OF VANCOMYCIN WITH 2-ACETOXY-D-PROPANOIC ACID | | Descriptor: | CHLORIDE ION, LACTIC ACID, VANCOMYCIN, ... | | Authors: | Loll, P.J, Kaplan, J, Selinsky, B, Axelsen, P.H. | | Deposit date: | 1999-07-20 | | Release date: | 1999-07-30 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Vancomycin binding to low-affinity ligands: delineating a minimum set of interactions necessary for high-affinity binding.
J.Med.Chem., 42, 1999
|
|
1C0R
 
 | | COMPLEX OF VANCOMYCIN WITH D-LACTIC ACID | | Descriptor: | CHLORIDE ION, LACTIC ACID, VANCOMYCIN, ... | | Authors: | Loll, P.J, Kaplan, J, Selinsky, B, Axelsen, P.H. | | Deposit date: | 1999-07-20 | | Release date: | 1999-07-30 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Vancomycin Binding to Low-Affinity Ligands: Delineating a Minimum Set of Interactions Necessary for High-Affinity Binding.
J.Med.Chem., 42, 1999
|
|
1C0T
 
 | | CRYSTAL STRUCTURE OF HIV-1 REVERSE TRANSCRIPTASE IN COMPLEX WITH BM+21.1326 | | Descriptor: | (R)-(+)9B-(3-METHYL)PHENYL-2,3-DIHYDROTHIAZOLO[2,3-A]ISOINDOL-5(9BH)-ONE, HIV-1 REVERSE TRANSCRIPTASE (A-CHAIN), HIV-1 REVERSE TRANSCRIPTASE (B-CHAIN) | | Authors: | Ren, J, Esnouf, R.M, Hopkins, A.L, Stuart, D.I, Stammers, D.K. | | Deposit date: | 1999-07-19 | | Release date: | 2000-07-19 | | Last modified: | 2014-11-12 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystallographic analysis of the binding modes of thiazoloisoindolinone non-nucleoside inhibitors to HIV-1 reverse transcriptase and comparison with modeling studies.
J.Med.Chem., 42, 1999
|
|
1C0U
 
 | | CRYSTAL STRUCTURE OF HIV-1 REVERSE TRANSCRIPTASE IN COMPLEX WITH BM+50.0934 | | Descriptor: | (R)-(+) 5(9BH)-OXO-9B-PHENYL-2,3-DIHYDROTHIAZOLO[2,3-A]ISOINDOL-3-CARBOXYLIC ACID METHYL ESTER, HIV-1 REVERSE TRANSCRIPTASE (A-CHAIN), HIV-1 REVERSE TRANSCRIPTASE (B-CHAIN) | | Authors: | Ren, J, Esnouf, R.M, Hopkins, A.L, Stuart, D.I, Stammers, D.K. | | Deposit date: | 1999-07-19 | | Release date: | 2000-07-19 | | Last modified: | 2014-11-12 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Crystallographic analysis of the binding modes of thiazoloisoindolinone non-nucleoside inhibitors to HIV-1 reverse transcriptase and comparison with modeling studies.
J.Med.Chem., 42, 1999
|
|
1C0V
 
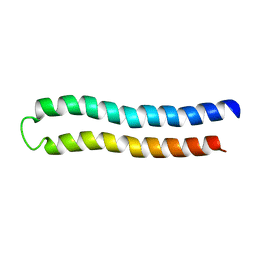 | | SUBUNIT C OF THE F1FO ATP SYNTHASE OF ESCHERICHIA COLI; NMR, 10 STRUCTURES | | Descriptor: | PROTEIN (F1FO ATPASE SUBUNIT C) | | Authors: | Girvin, M.E, Rastogi, V.K, Abildgaard, F, Markley, J.L, Fillingame, R.H. | | Deposit date: | 1999-07-22 | | Release date: | 1999-08-18 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the transmembrane H+-transporting subunit c of the F1F0 ATP synthase.
Biochemistry, 37, 1998
|
|
1C0W
 
 | | CRYSTAL STRUCTURE OF THE COBALT-ACTIVATED DIPHTHERIA TOXIN REPRESSOR-DNA COMPLEX REVEALS A METAL BINDING SH-LIKE DOMAIN | | Descriptor: | COBALT (II) ION, DIPHTHERIA TOXIN REPRESSOR, DNA (5'-D(P*AP*TP*TP*AP*GP*GP*TP*TP*AP*GP*CP*CP*TP*AP*CP*CP*CP*TP*AP*AP*T)-3'), ... | | Authors: | Pohl, E, Holmes, R.K, Hol, W.G. | | Deposit date: | 1999-07-22 | | Release date: | 2000-07-22 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structure of a cobalt-activated diphtheria toxin repressor-DNA complex reveals a metal-binding SH3-like domain.
J.Mol.Biol., 292, 1999
|
|
1C0Y
 
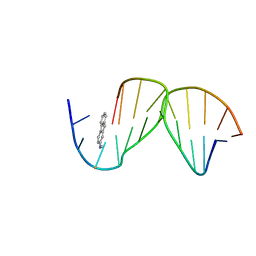 | | SOLUTION STRUCTURE OF THE [AF]-C8-DG ADDUCT POSITIONED OPPOSITE DA AT A TEMPLATE-PRIMER JUNCTION | | Descriptor: | 2-AMINOFLUORENE, DNA (5'-D(*AP*AP*CP*GP*CP*TP*AP*CP*CP*AP*TP*CP*C)-3'), DNA (5'-D(*GP*GP*AP*TP*GP*GP*TP*AP*GP*C)-3') | | Authors: | Gu, Z, Gorin, A, Hingerty, B.E, Broyde, S, Patel, D.J. | | Deposit date: | 1999-07-19 | | Release date: | 1999-08-31 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structures of aminofluorene [AF]-stacked conformers of the syn [AF]-C8-dG adduct positioned opposite dC or dA at a template-primer junction.
Biochemistry, 38, 1999
|
|
1C10
 
 | | CRYSTAL STRUCTURE OF HEW LYSOZYME UNDER PRESSURE OF XENON (8 BAR) | | Descriptor: | CHLORIDE ION, PROTEIN (LYSOZYME), SODIUM ION, ... | | Authors: | Prange, T, Schiltz, M, Pernot, L, Colloc'h, N, Longhi, S, Bourguet, W, Fourme, R. | | Deposit date: | 1999-07-16 | | Release date: | 1999-07-22 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Exploring hydrophobic sites in proteins with xenon or krypton.
Proteins, 30, 1998
|
|
1C11
 
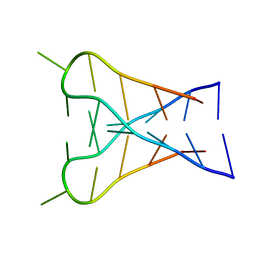 | | INTERCALATED D(TCCCGTTTCCA) DIMER, NMR, 7 STRUCTURES | | Descriptor: | DNA (5'-D(*TP*CP*CP*CP*GP*TP*TP*TP*CP*CP*A)-3') | | Authors: | Gallego, J, Chou, S.H, Reid, B.R. | | Deposit date: | 1998-07-15 | | Release date: | 1998-07-22 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Centromeric pyrimidine strands fold into an intercalated motif by forming a double hairpin with a novel T:G:G:T tetrad: solution structure of the d(TCCCGTTTCCA) dimer.
J.Mol.Biol., 273, 1997
|
|
1C12
 
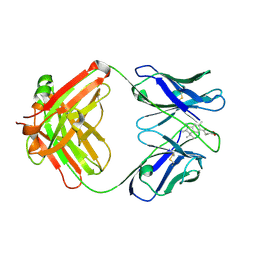 | | INSIGHT IN ODORANT PERCEPTION: THE CRYSTAL STRUCTURE AND BINDING CHARACTERISTICS OF ANTIBODY FRAGMENTS DIRECTED AGAINST THE MUSK ODORANT TRASEOLIDE | | Descriptor: | PROTEIN (ANTIBODY FRAGMENT FAB), TRAZEOLIDE | | Authors: | Langedijk, A.C, Spinelli, S, Anguille, C, Hermans, P, Nederlof, J, Butenandt, J, Honegger, A, Cambillau, C, Pluckthun, A. | | Deposit date: | 1999-07-20 | | Release date: | 1999-08-14 | | Last modified: | 2018-02-28 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Insight into odorant perception: the crystal structure and binding characteristics of antibody fragments directed against the musk odorant traseolide.
J.Mol.Biol., 292, 1999
|
|
1C14
 
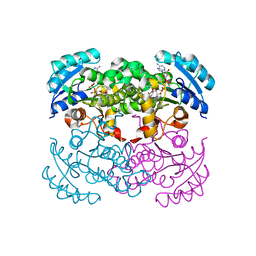 | | CRYSTAL STRUCTURE OF E COLI ENOYL REDUCTASE-NAD+-TRICLOSAN COMPLEX | | Descriptor: | ENOYL REDUCTASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, TRICLOSAN | | Authors: | Qiu, X, Janson, C, Court, R, Smyth, M, Payne, D, Abdel-Meguid, S. | | Deposit date: | 1999-07-20 | | Release date: | 2000-07-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Molecular basis for triclosan activity involves a flipping loop in the active site.
Protein Sci., 8, 1999
|
|
1C15
 
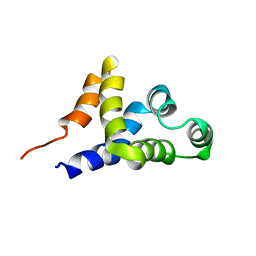 | | SOLUTION STRUCTURE OF APAF-1 CARD | | Descriptor: | APOPTOTIC PROTEASE ACTIVATING FACTOR 1 | | Authors: | Zhou, P, Chou, J, Olea, R.S, Yuan, J, Wagner, G. | | Deposit date: | 1999-07-20 | | Release date: | 1999-09-20 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of Apaf-1 CARD and its interaction with caspase-9 CARD: a structural basis for specific adaptor/caspase interaction.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
1C16
 
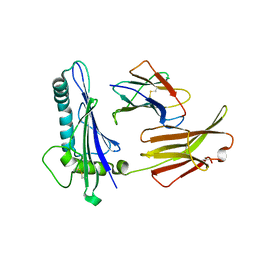 | | CRYSTAL STRUCTURE ANALYSIS OF THE GAMMA/DELTA T CELL LIGAND T22 | | Descriptor: | MHC-LIKE PROTEIN T22, PROTEIN (BETA-2-MICROGLOBULIN) | | Authors: | Wingren, C, Crowley, M.P, Degano, M, Chien, Y, Wilson, I.A. | | Deposit date: | 1999-07-20 | | Release date: | 2000-01-26 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crystal structure of a gammadelta T cell receptor ligand T22: a truncated MHC-like fold.
Science, 287, 2000
|
|
1C17
 
 | | A1C12 SUBCOMPLEX OF F1FO ATP SYNTHASE | | Descriptor: | ATP SYNTHASE SUBUNIT A, ATP SYNTHASE SUBUNIT C | | Authors: | Rastogi, V.K, Girvin, M.E. | | Deposit date: | 1999-07-20 | | Release date: | 1999-11-24 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural changes linked to proton translocation by subunit c of the ATP synthase.
Nature, 402, 1999
|
|
1C1A
 
 | |
1C1B
 
 | | CRYSTAL STRUCTURE OF HIV-1 REVERSE TRANSCRIPTASE IN COMPLEX WITH GCA-186 | | Descriptor: | 6-(3',5'-DIMETHYLBENZYL)-1-ETHOXYMETHYL-5-ISOPROPYLURACIL, HIV-1 REVERSE TRANSCRIPTASE (A-CHAIN), HIV-1 REVERSE TRANSCRIPTASE (B-CHAIN) | | Authors: | Hopkins, A.L, Ren, J, Tanaka, H, Baba, B, Okamato, M, Stuart, D.I, Stammers, D.K. | | Deposit date: | 1999-07-21 | | Release date: | 2000-07-21 | | Last modified: | 2014-11-12 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Design of MKC-442 (emivirine) analogues with improved activity against drug-resistant HIV mutants.
J.Med.Chem., 42, 1999
|
|
1C1C
 
 | | CRYSTAL STRUCTURE OF HIV-1 REVERSE TRANSCRIPTASE IN COMPLEX WITH TNK-6123 | | Descriptor: | 6-(cyclohexylsulfanyl)-1-(ethoxymethyl)-5-(1-methylethyl)pyrimidine-2,4(1H,3H)-dione, HIV-1 REVERSE TRANSCRIPTASE (A-CHAIN), HIV-1 REVERSE TRANSCRIPTASE (B-CHAIN) | | Authors: | Hopkins, A.L, Ren, J, Tanaka, H, Baba, M, Okamato, M, Stuart, D.I, Stammers, D.K. | | Deposit date: | 1999-07-21 | | Release date: | 2000-07-21 | | Last modified: | 2014-11-12 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Design of MKC-442 (emivirine) analogues with improved activity against drug-resistant HIV mutants.
J.Med.Chem., 42, 1999
|
|
1C1D
 
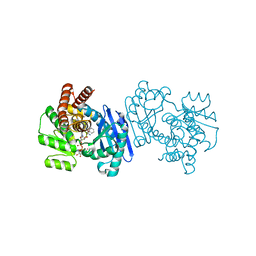 | | L-PHENYLALANINE DEHYDROGENASE STRUCTURE IN TERNARY COMPLEX WITH NADH AND L-PHENYLALANINE | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, ISOPROPYL ALCOHOL, L-PHENYLALANINE DEHYDROGENASE, ... | | Authors: | Vanhooke, J.L, Thoden, J.B. | | Deposit date: | 1999-07-21 | | Release date: | 2000-08-30 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Rhodococcus L-phenylalanine dehydrogenase: kinetics, mechanism, and structural basis for catalytic specificity.
Biochemistry, 39, 2000
|
|
1C1E
 
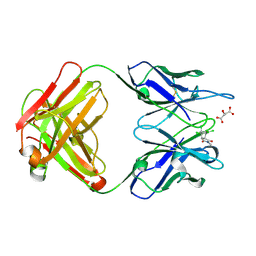 | | CRYSTAL STRUCTURE OF A DIELS-ALDERASE CATALYTIC ANTIBODY 1E9 IN COMPLEX WITH ITS HAPTEN | | Descriptor: | 1,7,8,9,10,10-HEXACHLORO-4-METHYL-4-AZA-TRICYCLO[5.2.1.0(2,6)]DEC-8-ENE-3,5-DIONE, CATALYTIC ANTIBODY 1E9 (HEAVY CHAIN), CATALYTIC ANTIBODY 1E9 (LIGHT CHAIN), ... | | Authors: | Xu, J, Wilson, I.A. | | Deposit date: | 1999-07-22 | | Release date: | 2000-03-01 | | Last modified: | 2013-01-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Evolution of shape complementarity and catalytic efficiency from a primordial antibody template.
Science, 286, 1999
|
|
1C1F
 
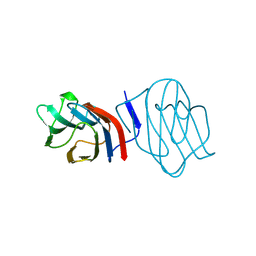 | | LIGAND-FREE CONGERIN I | | Descriptor: | PROTEIN (CONGERIN I) | | Authors: | Shirai, T, Mitsuyama, C, Niwa, Y, Matsui, Y, Hotta, H, Yamane, T, Kamiya, H, Ishii, C, Ogawa, T, Muramoto, K. | | Deposit date: | 1999-03-03 | | Release date: | 1999-10-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | High-resolution structure of the conger eel galectin, congerin I, in lactose-liganded and ligand-free forms: emergence of a new structure class by accelerated evolution.
Structure Fold.Des., 7, 1999
|
|
1C1G
 
 | |
