5TV5
 
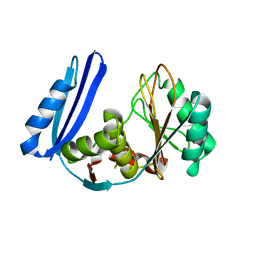 | | BioW from Aquifex aeoulicus | | Descriptor: | 6-carboxyhexanoate--CoA ligase | | Authors: | Estrada, P, Manandhar, M, Dong, S.-H, Deveryshetty, J, Agarwal, V, Cronan, J.E, Nair, S.K. | | Deposit date: | 2016-11-08 | | Release date: | 2016-12-07 | | Last modified: | 2017-05-31 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The pimeloyl-CoA synthetase BioW defines a new fold for adenylate-forming enzymes.
Nat. Chem. Biol., 13, 2017
|
|
5TV8
 
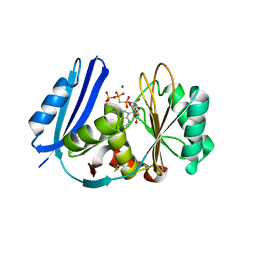 | | A. aeolicus BioW with AMP-CPP and pimelate | | Descriptor: | 6-carboxyhexanoate--CoA ligase, DIPHOSPHOMETHYLPHOSPHONIC ACID ADENOSYL ESTER, MAGNESIUM ION, ... | | Authors: | Estrada, P, Manandhar, M, Dong, S.-H, Deveryshetty, J, Agarwal, V, Cronan, J.E, Nair, S.K. | | Deposit date: | 2016-11-08 | | Release date: | 2016-12-07 | | Last modified: | 2017-05-31 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | The pimeloyl-CoA synthetase BioW defines a new fold for adenylate-forming enzymes.
Nat. Chem. Biol., 13, 2017
|
|
2YX5
 
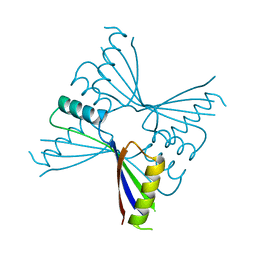 | | Crystal Structure of Methanocaldococcus jannaschii PurS, One of the Subunits of Formylglycinamide Ribonucleotide Amidotransferase in the Purine Biosynthetic Pathway | | Descriptor: | UPF0062 protein MJ1593 | | Authors: | Kanagawa, M, Baba, S, Agari, Y, Chen, L.Q, Fu, Z.-Q, Chrzas, J, Wang, B.C, Kuramitsu, S, Yokoyama, S, Kawai, G, Sampei, G, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-24 | | Release date: | 2007-10-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of Methanocaldococcus jannaschii PurS, One of the Subunits of Formylglycinamide Ribonucleotide Amidotransferase in the Purine Biosynthetic Pathway
To be Published
|
|
4Z7Y
 
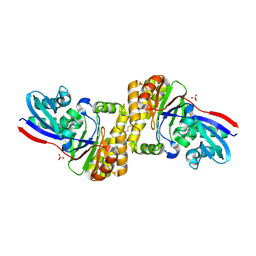 | | diphosphomevalonate decarboxylase from the Sulfolobus solfataricus, space group P21 | | Descriptor: | Diphosphomevalonate decarboxylase, SULFATE ION | | Authors: | Hattori, A, Unno, H, Hemmi, H. | | Deposit date: | 2015-04-08 | | Release date: | 2015-09-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | In Vivo Formation of the Protein Disulfide Bond That Enhances the Thermostability of Diphosphomevalonate Decarboxylase, an Intracellular Enzyme from the Hyperthermophilic Archaeon Sulfolobus solfataricus
J.Bacteriol., 197, 2015
|
|
2YWV
 
 | | Crystal structure of SAICAR synthetase from Geobacillus kaustophilus | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Phosphoribosylaminoimidazole succinocarboxamide synthetase, ... | | Authors: | Kanagawa, M, Baba, S, Kuramitsu, S, Yokoyama, S, Kawai, G, Sampei, G, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-23 | | Release date: | 2007-10-23 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of SAICAR synthetase from Geobacillus kaustophilus
To be Published
|
|
2X9N
 
 | | High resolution structure of TbPTR1 in complex with cyromazine | | Descriptor: | (4S,5S)-1,2-DITHIANE-4,5-DIOL, ACETATE ION, DITHIANE DIOL, ... | | Authors: | Dawson, A, Tulloch, L.B, Barrack, K.L, Hunter, W.N. | | Deposit date: | 2010-03-23 | | Release date: | 2010-06-30 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | High-Resolution Structures of Trypanosoma Brucei Pteridine Reductase Ligand Complexes Inform on the Placement of New Molecular Entities in the Active Site of a Potential Drug Target
Acta Crystallogr.,Sect.D, 66, 2010
|
|
4Z7C
 
 | | Diphosphomevalonate decarboxylase from the Sulfolobus solfataricus, space group h32 | | Descriptor: | Diphosphomevalonate decarboxylase, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Unno, H, Hattori, A, Hemmi, H. | | Deposit date: | 2015-04-07 | | Release date: | 2015-09-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | In Vivo Formation of the Protein Disulfide Bond That Enhances the Thermostability of Diphosphomevalonate Decarboxylase, an Intracellular Enzyme from the Hyperthermophilic Archaeon Sulfolobus solfataricus
J.Bacteriol., 197, 2015
|
|
4JA0
 
 | | Crystal structure of the invertebrate bi-functional purine biosynthesis enzyme PAICS at 2.8 A resolution | | Descriptor: | Phosphoribosylaminoimidazole carboxylase, SULFATE ION | | Authors: | Taschner, M, Basquin, J, Benda, C, Lorentzen, E. | | Deposit date: | 2013-02-18 | | Release date: | 2013-02-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of the invertebrate bifunctional purine biosynthesis enzyme PAICS at 2.8 angstrom resolution.
Proteins, 81, 2013
|
|
4N0H
 
 | | Crystal structure of S. cerevisiae mitochondrial GatFAB | | Descriptor: | Glutamyl-tRNA(Gln) amidotransferase subunit A, mitochondrial, Glutamyl-tRNA(Gln) amidotransferase subunit B, ... | | Authors: | Araiso, Y, Ishitani, R, Nureki, O. | | Deposit date: | 2013-10-02 | | Release date: | 2014-04-16 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.952 Å) | | Cite: | Crystal structure of Saccharomyces cerevisiae mitochondrial GatFAB reveals a novel subunit assembly in tRNA-dependent amidotransferases
Nucleic Acids Res., 42, 2014
|
|
3LES
 
 | | 2F5 Epitope scaffold ES2 | | Descriptor: | RNA polymerase sigma factor, SULFATE ION | | Authors: | Ofek, G, Guenaga, F.J, Schief, W.R, Skinner, J, Wyatt, R, Baker, D, Kwong, P.D. | | Deposit date: | 2010-01-15 | | Release date: | 2010-09-29 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.77 Å) | | Cite: | Elicitation of structure-specific antibodies by epitope scaffolds.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
4CRJ
 
 | | Staphylococcus aureus 7,8-Dihydro-6-hydroxymethylpterin- pyrophosphokinase in complex with AMPCPP and an inhibitor | | Descriptor: | 2-amino-8-{[2-(4-methoxyphenyl)-2-oxoethyl]sulfanyl}-1,9-dihydro-6H-purin-6-one, 7,8-DIHYDRO-6-HYDROXYMETHYLPTERIN-PYROPHOSPHOKINASE (HPPK), DIPHOSPHOMETHYLPHOSPHONIC ACID ADENOSYL ESTER, ... | | Authors: | Dennis, M.L, Swarbrick, J.D, Peat, T.S. | | Deposit date: | 2014-02-27 | | Release date: | 2015-01-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-Based Design and Development of Functionalized Mercaptoguanine Derivatives as Inhibitors of the Folate Biosynthesis Pathway Enzyme 6-Hydroxymethyl-7,8-Dihydropterin Pyrophosphokinase from Staphylococcus Aureus.
J.Med.Chem., 57, 2014
|
|
1PX5
 
 | | Crystal structure of the 2'-specific and double-stranded RNA-activated interferon-induced antiviral protein 2'-5'-oligoadenylate synthetase | | Descriptor: | 2'-5'-oligoadenylate synthetase 1, SULFATE ION | | Authors: | Hartmann, R, Justesen, J, Sarkar, S.N, Sen, G.C, Yee, V.C. | | Deposit date: | 2003-07-02 | | Release date: | 2003-11-25 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Crystal structure of the 2'-specific and double-stranded RNA-activated interferon-induced antiviral protein 2'-5'-oligoadenylate synthetase
Mol.Cell, 12, 2003
|
|
4N0I
 
 | | Crystal structure of S. cerevisiae mitochondrial GatFAB in complex with glutamine | | Descriptor: | GLUTAMINE, Glutamyl-tRNA(Gln) amidotransferase subunit A, mitochondrial, ... | | Authors: | Araiso, Y, Ishitani, R, Nureki, O. | | Deposit date: | 2013-10-02 | | Release date: | 2014-04-16 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | Crystal structure of Saccharomyces cerevisiae mitochondrial GatFAB reveals a novel subunit assembly in tRNA-dependent amidotransferases
Nucleic Acids Res., 42, 2014
|
|
4CWB
 
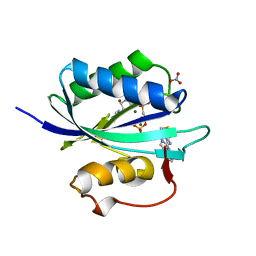 | | Staphylococcus aureus 7,8-Dihydro-6-hydroxymethylpterin- pyrophosphokinase in complex with AMPCPP and an inhibitor | | Descriptor: | 2-amino-8-[2-oxo-2-(4-phenylphenyl)ethyl]sulfanyl-1,9-dihydropurin-6-one, 7,8-DIHYDRO-6-HYDROXYMETHYLPTERIN-PYROPHOSPHOKINASE, DIPHOSPHOMETHYLPHOSPHONIC ACID ADENOSYL ESTER, ... | | Authors: | Dennis, M.L, Swarbrick, J.D, Peat, T.S. | | Deposit date: | 2014-04-02 | | Release date: | 2015-01-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Structure-Based Design and Development of Functionalized Mercaptoguanine Derivatives as Inhibitors of the Folate Biosynthesis Pathway Enzyme 6-Hydroxymethyl-7,8-Dihydropterin Pyrophosphokinase from Staphylococcus Aureus.
J.Med.Chem., 57, 2014
|
|
4CYU
 
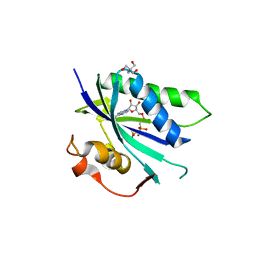 | | Staphylococcus aureus 7,8-Dihydro-6-hydroxymethylpterin- pyrophosphokinase in complex with AMPCPP | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 7,8-DIHYDRO-6-HYDROXYMETHYLPTERIN-PYROPHOSPHOKINASE, DIPHOSPHOMETHYLPHOSPHONIC ACID ADENOSYL ESTER, ... | | Authors: | Dennis, M.L, Swarbrick, J.D, Peat, T.S. | | Deposit date: | 2014-04-15 | | Release date: | 2015-01-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure-Based Design and Development of Functionalized Mercaptoguanine Derivatives as Inhibitors of the Folate Biosynthesis Pathway Enzyme 6-Hydroxymethyl-7,8-Dihydropterin Pyrophosphokinase from Staphylococcus Aureus.
J.Med.Chem., 57, 2014
|
|
4RCN
 
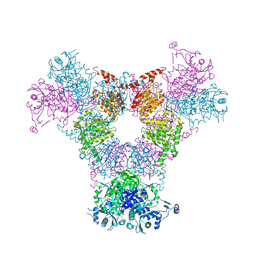 | |
2CUW
 
 | | Crystal Structure of Thermus thermophilus PurS, one of the subunits of Formylglycinamide Ribonucleotide Amidotransferase in the purine biosynthetic pathway | | Descriptor: | PurS | | Authors: | Yanai, H, Kanagawa, M, Sampei, G, Kawai, G, Yokoyama, S, Kuramitsu, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-05-30 | | Release date: | 2005-11-30 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of Thermus thermophilus PurS, One of the Subunits of Formylglycinamide Ribonucleotide Amidotransferase in the Purine Biosynthetic Pathway
To be Published
|
|
2OZX
 
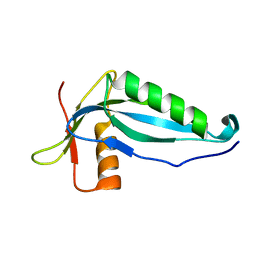 | |
2EQA
 
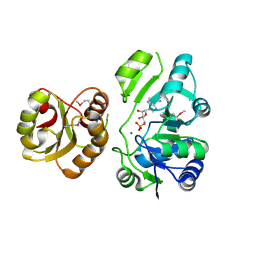 | | Crystal Structure of the hypothetical Sua5 protein from Sulfolobus tokodaii | | Descriptor: | ADENOSINE MONOPHOSPHATE, Hypothetical protein ST1526, MAGNESIUM ION | | Authors: | Agari, Y, Shinkai, A, Yokoyama, S, Kuramitsu, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-03-30 | | Release date: | 2008-01-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-ray crystal structure of a hypothetical Sua5 protein from Sulfolobus tokodaii strain 7
Proteins, 70, 2008
|
|
2OZW
 
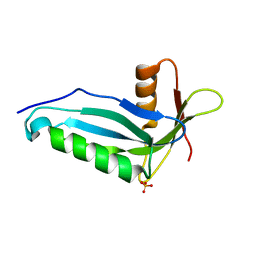 | |
2GI3
 
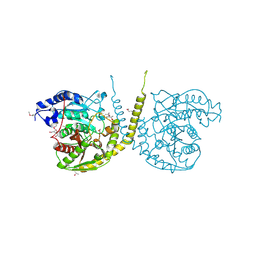 | |
2D6F
 
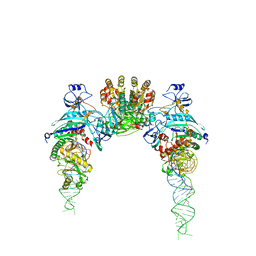 | |
2CQY
 
 | |
2DGB
 
 | |
3O3N
 
 | | (R)-2-hydroxyisocaproyl-CoA dehydratase in complex with its substrate (R)-2-hydroxyisocaproyl-CoA | | Descriptor: | HYDROSULFURIC ACID, IRON/SULFUR CLUSTER, S-[2-[3-[[(2R)-4-[[[(2R,3S,4R,5R)-5-(6-aminopurin-9-yl)-4-hydroxy-3-phosphonooxy-oxolan-2-yl]methoxy-hydroxy-phosphoryl ]oxy-hydroxy-phosphoryl]oxy-2-hydroxy-3,3-dimethyl-butanoyl]amino]propanoylamino]ethyl] (2R)-2-hydroxy-4-methyl-pentanethioate, ... | | Authors: | Knauer, S.H, Buckel, W, Dobbek, H. | | Deposit date: | 2010-07-25 | | Release date: | 2011-03-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Basis for Reductive Radical Formation and Electron Recycling in (R)-2-Hydroxyisocaproyl-CoA Dehydratase.
J.Am.Chem.Soc., 133, 2011
|
|
