8RQB
 
 | | Cryo-EM structure of mouse heavy-chain apoferritin | | Descriptor: | FE (III) ION, Ferritin heavy chain, N-terminally processed, ... | | Authors: | Nazarov, S, Myasnikov, A, Stahlberg, H. | | Deposit date: | 2024-01-17 | | Release date: | 2024-03-13 | | Last modified: | 2024-10-02 | | Method: | ELECTRON MICROSCOPY (1.09 Å) | | Cite: | Low-dose cryo-electron ptychography of proteins at sub-nanometer resolution.
Nat Commun, 15, 2024
|
|
3PYP
 
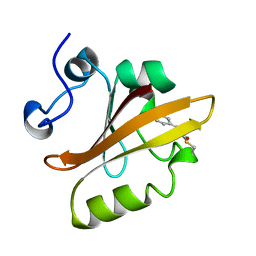 | | PHOTOACTIVE YELLOW PROTEIN, CRYOTRAPPED EARLY LIGHT CYCLE INTERMEDIATE | | Descriptor: | 4'-HYDROXYCINNAMIC ACID, PHOTOACTIVE YELLOW PROTEIN | | Authors: | Genick, U.K, Soltis, S.M, Kuhn, P, Canestrelli, I.L, Getzoff, E.D. | | Deposit date: | 1998-07-28 | | Release date: | 1999-06-01 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (0.85 Å) | | Cite: | Structure at 0.85 A resolution of an early protein photocycle intermediate.
Nature, 392, 1998
|
|
6IS6
 
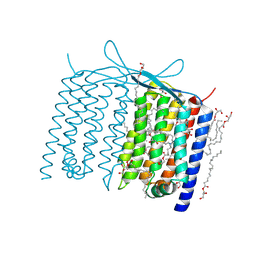 | | Crystal structure of Thermoplasmatales archaeon heliorhodopsin | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, RETINAL, heliorhodopsin | | Authors: | Shihoya, W, Yamashita, K, Nureki, O. | | Deposit date: | 2018-11-15 | | Release date: | 2019-09-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of heliorhodopsin.
Nature, 574, 2019
|
|
6JYA
 
 | | Structure of dark-state marine bacterial chloride importer, NM-R3, with CW laser (ND-10%) at 95K. | | Descriptor: | CHLORIDE ION, Chloride pumping rhodopsin, OLEIC ACID, ... | | Authors: | Yun, J.H, Ohki, M, Park, S.Y, Lee, W. | | Deposit date: | 2019-04-26 | | Release date: | 2020-03-04 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.803 Å) | | Cite: | Pumping mechanism of NM-R3, a light-driven bacterial chloride importer in the rhodopsin family.
Sci Adv, 6, 2020
|
|
6JYF
 
 | | Structure of light-state marine bacterial chloride importer, NM-R3, with Pulse laser (ND-1%) at 140K. | | Descriptor: | CHLORIDE ION, Chloride pumping rhodopsin, OLEIC ACID, ... | | Authors: | Yun, J.H, Ohki, M, Park, S.Y, Lee, W. | | Deposit date: | 2019-04-26 | | Release date: | 2020-03-04 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.004 Å) | | Cite: | Pumping mechanism of NM-R3, a light-driven bacterial chloride importer in the rhodopsin family.
Sci Adv, 6, 2020
|
|
6JYE
 
 | | Structure of dark-state marine bacterial chloride importer, NM-R3, with Pulse laser (ND-1%) at 140K. | | Descriptor: | CHLORIDE ION, Chloride pumping rhodopsin, OLEIC ACID, ... | | Authors: | Yun, J.H, Ohki, M, Park, S.Y, Lee, W. | | Deposit date: | 2019-04-26 | | Release date: | 2020-03-04 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Pumping mechanism of NM-R3, a light-driven bacterial chloride importer in the rhodopsin family.
Sci Adv, 6, 2020
|
|
6JY6
 
 | | Structure of dark-state marine bacterial chloride importer, NM-R3, with Pulse laser (ND-1%) at 95K. | | Descriptor: | CHLORIDE ION, Chloride pumping rhodopsin, OLEIC ACID, ... | | Authors: | Yun, J.H, Ohki, M, Park, S.Y, Lee, W. | | Deposit date: | 2019-04-26 | | Release date: | 2020-03-04 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Pumping mechanism of NM-R3, a light-driven bacterial chloride importer in the rhodopsin family.
Sci Adv, 6, 2020
|
|
6JYC
 
 | | Structure of dark-state marine bacterial chloride importer, NM-R3, with CW laser (ND-30%) at 95K. | | Descriptor: | CHLORIDE ION, Chloride pumping rhodopsin, OLEIC ACID, ... | | Authors: | Yun, J.H, Ohki, M, Park, S.Y, Lee, W. | | Deposit date: | 2019-04-26 | | Release date: | 2020-03-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.892 Å) | | Cite: | Pumping mechanism of NM-R3, a light-driven bacterial chloride importer in the rhodopsin family.
Sci Adv, 6, 2020
|
|
6JY8
 
 | | Structure of dark-state marine bacterial chloride importer, NM-R3, with CW laser (ND-3%) at 95K. | | Descriptor: | CHLORIDE ION, Chloride pumping rhodopsin, OLEIC ACID, ... | | Authors: | Yun, J.H, Ohki, M, Park, S.Y, Lee, W. | | Deposit date: | 2019-04-26 | | Release date: | 2020-03-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Pumping mechanism of NM-R3, a light-driven bacterial chloride importer in the rhodopsin family.
Sci Adv, 6, 2020
|
|
6JY7
 
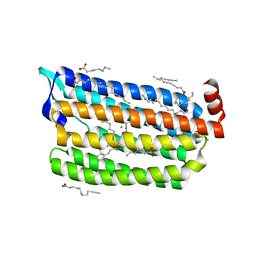 | | Structure of light-state marine bacterial chloride importer, NM-R3, with Pulse laser (ND-1%) at 95K. | | Descriptor: | CHLORIDE ION, Chloride pumping rhodopsin, OLEIC ACID, ... | | Authors: | Yun, J.H, Ohki, M, Park, S.Y, Lee, W. | | Deposit date: | 2019-04-26 | | Release date: | 2020-03-04 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Pumping mechanism of NM-R3, a light-driven bacterial chloride importer in the rhodopsin family.
Sci Adv, 6, 2020
|
|
6JYD
 
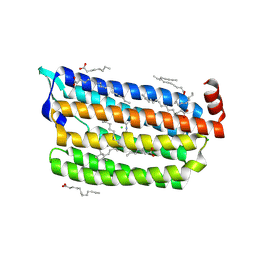 | | Structure of light-state marine bacterial chloride importer, NM-R3, with CW laser (ND-30%) at 95K. | | Descriptor: | CHLORIDE ION, Chloride pumping rhodopsin, OLEIC ACID, ... | | Authors: | Yun, J.H, Ohki, M, Park, S.Y, Lee, W. | | Deposit date: | 2019-04-26 | | Release date: | 2020-03-04 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.007 Å) | | Cite: | Pumping mechanism of NM-R3, a light-driven bacterial chloride importer in the rhodopsin family.
Sci Adv, 6, 2020
|
|
6JY9
 
 | | Structure of light-state marine bacterial chloride importer, NM-R3, with CW laser (ND-3%) at 95K. | | Descriptor: | CHLORIDE ION, Chloride pumping rhodopsin, OLEIC ACID, ... | | Authors: | Yun, J.H, Ohki, M, Park, S.Y, Lee, W. | | Deposit date: | 2019-04-26 | | Release date: | 2020-03-04 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Pumping mechanism of NM-R3, a light-driven bacterial chloride importer in the rhodopsin family.
Sci Adv, 6, 2020
|
|
6JYB
 
 | | Structure of light-state marine bacterial chloride importer, NM-R3, with CW laser (ND-10%) at 95K. | | Descriptor: | CHLORIDE ION, Chloride pumping rhodopsin, OLEIC ACID, ... | | Authors: | Yun, J.H, Ohki, M, Park, S.Y, Lee, W. | | Deposit date: | 2019-04-26 | | Release date: | 2020-03-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.802 Å) | | Cite: | Pumping mechanism of NM-R3, a light-driven bacterial chloride importer in the rhodopsin family.
Sci Adv, 6, 2020
|
|
1UAZ
 
 | |
6WP8
 
 | | Proton-pumping mutant of Mastigocladopsis repens rhodopsin chloride pump | | Descriptor: | Proton-pumping rhodopsin chloride pump, RETINAL, octyl beta-D-glucopyranoside | | Authors: | Besaw, J.E, Ernst, O.P, Ou, W, Morizumi, T. | | Deposit date: | 2020-04-26 | | Release date: | 2020-07-29 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The crystal structures of a chloride-pumping microbial rhodopsin and its proton-pumping mutant illuminate proton transfer determinants.
J.Biol.Chem., 295, 2020
|
|
6XL3
 
 | | Mastigocladopsis repens rhodopsin chloride pump | | Descriptor: | CHLORIDE ION, DECANE, Mastigocladopsis repens rhodopsin chloride pump, ... | | Authors: | Besaw, J.E, Ernst, O.P, Ou, W, Morizumi, T. | | Deposit date: | 2020-06-28 | | Release date: | 2020-07-29 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | The crystal structures of a chloride-pumping microbial rhodopsin and its proton-pumping mutant illuminate proton transfer determinants.
J.Biol.Chem., 295, 2020
|
|
7E4G
 
 | | Crystal structure of schizorhodopsin 4 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, RETINAL, SULFATE ION, ... | | Authors: | Shihoya, W, Nureki, O. | | Deposit date: | 2021-02-12 | | Release date: | 2021-02-24 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of schizorhodopsin reveals mechanism of inward proton pumping.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7LHJ
 
 | |
7LHN
 
 | |
7LHM
 
 | |
7LHO
 
 | |
2M06
 
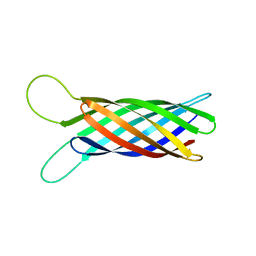 | | NMR structure of OmpX in phopspholipid nanodiscs | | Descriptor: | Outer membrane protein X | | Authors: | Hagn, F.X, Etzkorn, M, Raschle, T, Wagner, G, Membrane Protein Structures by Solution NMR (MPSbyNMR) | | Deposit date: | 2012-10-21 | | Release date: | 2012-12-12 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Optimized phospholipid bilayer nanodiscs facilitate high-resolution structure determination of membrane proteins.
J.Am.Chem.Soc., 135, 2013
|
|
2M07
 
 | | NMR structure of OmpX in DPC micelles | | Descriptor: | Outer membrane protein X | | Authors: | Hagn, F.X, Etzkorn, M, Raschle, T, Wagner, G, Membrane Protein Structures by Solution NMR (MPSbyNMR) | | Deposit date: | 2012-10-21 | | Release date: | 2012-12-12 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Optimized phospholipid bilayer nanodiscs facilitate high-resolution structure determination of membrane proteins.
J.Am.Chem.Soc., 135, 2013
|
|
5KKI
 
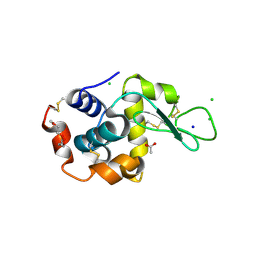 | | 1.7-Angstrom in situ Mylar structure of hen egg-white lysozyme (HEWL) at 100 K | | Descriptor: | 3,6,9,12,15,18,21,24-OCTAOXAHEXACOSAN-1-OL, ACETIC ACID, CHLORIDE ION, ... | | Authors: | Broecker, J, Ernst, O.P. | | Deposit date: | 2016-06-21 | | Release date: | 2017-02-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A Versatile System for High-Throughput In Situ X-ray Screening and Data Collection of Soluble and Membrane-Protein Crystals.
Cryst Growth Des, 16, 2016
|
|
5KKJ
 
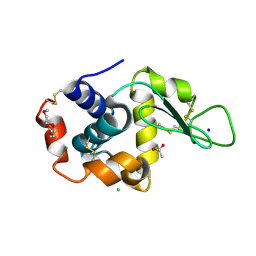 | | 2.0-Angstrom In situ Mylar structure of hen egg-white lysozyme (HEWL) at 293 K | | Descriptor: | 3,6,9,12,15,18,21,24-OCTAOXAHEXACOSAN-1-OL, ACETIC ACID, CHLORIDE ION, ... | | Authors: | Broecker, J, Ernst, O.P. | | Deposit date: | 2016-06-21 | | Release date: | 2017-02-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | A Versatile System for High-Throughput In Situ X-ray Screening and Data Collection of Soluble and Membrane-Protein Crystals.
Cryst Growth Des, 16, 2016
|
|
