2Z8X
 
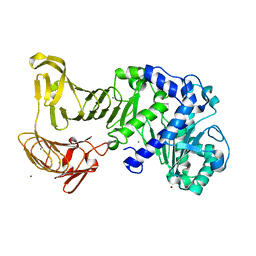 | | Crystal structure of extracellular lipase from Pseudomonas sp. MIS38 | | Descriptor: | CALCIUM ION, Lipase, ZINC ION | | Authors: | Angkawidjaja, C, You, D.J, Matsumura, H, Koga, Y, Takano, K, Kanaya, S. | | Deposit date: | 2007-09-11 | | Release date: | 2007-10-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Crystal structure of a family I.3 lipase from Pseudomonas sp. MIS38 in a closed conformation
FEBS Lett., 581, 2007
|
|
2Z8Z
 
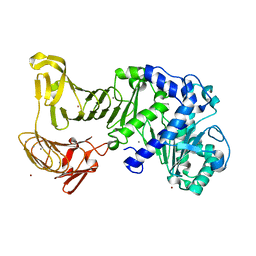 | | Crystal structure of a platinum-bound S445C mutant of Pseudomonas sp. MIS38 lipase | | Descriptor: | CALCIUM ION, Lipase, PLATINUM (II) ION, ... | | Authors: | Angkawidjaja, C, You, D.J, Matsumura, H, Koga, Y, Takano, K, Kanaya, S. | | Deposit date: | 2007-09-13 | | Release date: | 2007-10-30 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of a family I.3 lipase from Pseudomonas sp. MIS38 in a closed conformation
FEBS Lett., 581, 2007
|
|
3A70
 
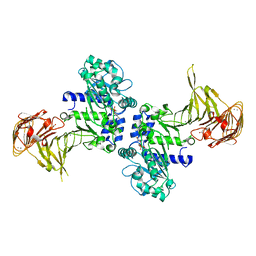 | | Crystal structure of Pseudomonas sp. MIS38 lipase in complex with diethyl phosphate | | Descriptor: | ACETATE ION, CALCIUM ION, DIETHYL PHOSPHONATE, ... | | Authors: | Angkawidjaja, C, Matsumura, H, Koga, Y, Takano, K, Kanaya, S. | | Deposit date: | 2009-09-10 | | Release date: | 2010-05-26 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | X-ray Crystallographic and MD Simulation Studies on the Mechanism of Interfacial Activation of a Family I.3 Lipase with Two Lids
J.Mol.Biol., 2010
|
|
3A6Z
 
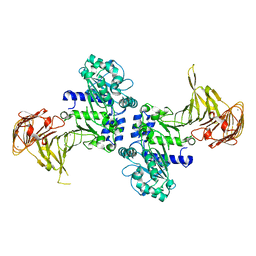 | | Crystal structure of Pseudomonas sp. MIS38 lipase (PML) in the open conformation following dialysis against Ca-free buffer | | Descriptor: | CALCIUM ION, Lipase | | Authors: | Angkawidjaja, C, Matsumura, H, Koga, Y, Takano, K, Kanaya, S. | | Deposit date: | 2009-09-10 | | Release date: | 2010-05-26 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | X-ray Crystallographic and MD Simulation Studies on the Mechanism of Interfacial Activation of a Family I.3 Lipase with Two Lids
J.Mol.Biol., 2010
|
|
2ZVD
 
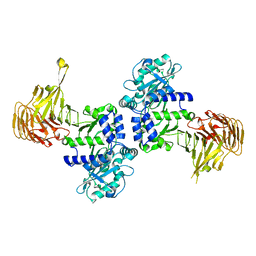 | | Crystal structure of Pseudomonas sp. MIS38 lipase in an open conformation | | Descriptor: | CALCIUM ION, Lipase | | Authors: | Angkawidjaja, C, Matsumura, H, Koga, Y, Takano, K, Kanaya, S. | | Deposit date: | 2008-11-05 | | Release date: | 2009-11-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | X-ray Crystallographic and MD Simulation Studies on the Mechanism of Interfacial Activation of a Family I.3 Lipase with Two Lids
J.Mol.Biol., 2010
|
|
2ZJ7
 
 | |
2ZJ6
 
 | |
