1IWX
 
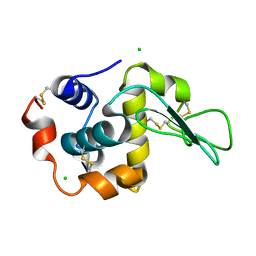 | | Crystal Structure Analysis of Human lysozyme at 161K. | | Descriptor: | CHLORIDE ION, LYSOZYME C | | Authors: | Joti, Y, Nakasako, M, Kidera, A, Go, N. | | Deposit date: | 2002-06-03 | | Release date: | 2002-09-04 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Nonlinear temperature dependence of the crystal structure of lysozyme: correlation between coordinate shifts and thermal factors.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
1IWY
 
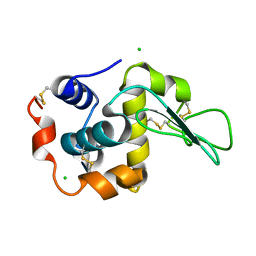 | | Crystal Structure Analysis of Human lysozyme at 170K. | | Descriptor: | CHLORIDE ION, LYSOZYME C | | Authors: | Joti, Y, Nakasako, M, Kidera, A, Go, N. | | Deposit date: | 2002-06-03 | | Release date: | 2002-09-04 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Nonlinear temperature dependence of the crystal structure of lysozyme: correlation between coordinate shifts and thermal factors.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
1IWZ
 
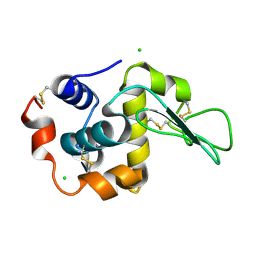 | | Crystal Structure Analysis of Human lysozyme at 178K. | | Descriptor: | CHLORIDE ION, LYSOZYME C | | Authors: | Joti, Y, Nakasako, M, Kidera, A, Go, N. | | Deposit date: | 2002-06-03 | | Release date: | 2002-09-04 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Nonlinear temperature dependence of the crystal structure of lysozyme: correlation between coordinate shifts and thermal factors.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
1IX0
 
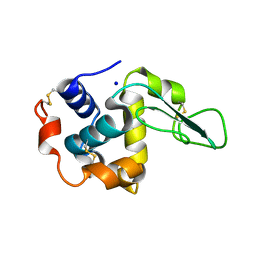 | | I59A-3SS human lysozyme | | Descriptor: | SODIUM ION, lysozyme | | Authors: | Takano, K, Yamagata, Y, Yutani, K. | | Deposit date: | 2002-06-06 | | Release date: | 2003-07-22 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Buried water molecules contribute to the conformational stability of a protein
PROTEIN ENG., 16, 2003
|
|
1IX1
 
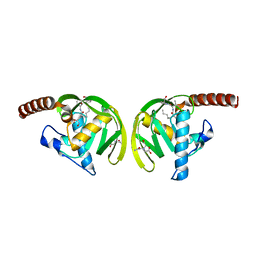 | | Crystal Structure of P.aeruginosa Peptide deformylase Complexed with Antibiotic Actinonin | | Descriptor: | (CARBAMOYLMETHYL-CARBOXYMETHYL-AMINO)-ACETIC ACID, ACTINONIN, ZINC ION, ... | | Authors: | Kim, H.-W, Yoon, H.-J, Lee, J.Y, Han, B.W, Yang, J.K, Lee, B.I, Ahn, H.J, Lee, H.H, Suh, S.W. | | Deposit date: | 2002-06-07 | | Release date: | 2003-09-09 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of peptide deformylase from Staphylococcus aureus in complex with actinonin, a naturally occurring antibacterial agent
Proteins, 57, 2004
|
|
1IX2
 
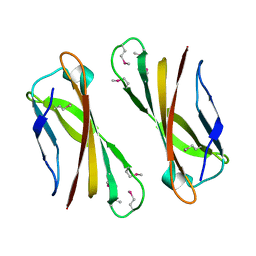 | | Crystal Structure of Selenomethionine PcoC, a Copper Resistance Protein from Escherichia coli | | Descriptor: | PcoC copper resistance protein | | Authors: | Wernimont, A.K, Huffman, D.L, Finney, L.A, Demeler, B, O'Halloran, T.V, Rosenzweig, A.C. | | Deposit date: | 2002-06-10 | | Release date: | 2002-11-27 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structure and dimerization equilibria of PcoC, a methionine-rich copper resistance protein from Escherichia coli
J.BIOL.INORG.CHEM., 8, 2003
|
|
1IX3
 
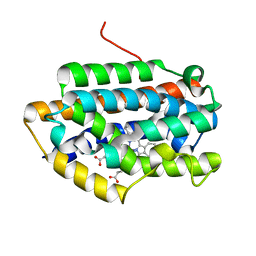 | | Crystal Structure of Rat Heme Oxygenase-1 in complex with Heme bound to Cyanide | | Descriptor: | CYANIDE ION, HEME OXYGENASE-1, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Sugishima, M, Sakamoto, H, Omata, Y, Hayashi, S, Noguchi, M, Fukuyama, K. | | Deposit date: | 2002-06-10 | | Release date: | 2003-09-02 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structures of Ferrous and CO-, CN(-)-, and NO-Bound Forms of Rat Heme Oxygenase-1 (HO-1) in Complex with Heme: Structural Implications for Discrimination between CO and O(2) in HO-1.
Biochemistry, 42, 2003
|
|
1IX4
 
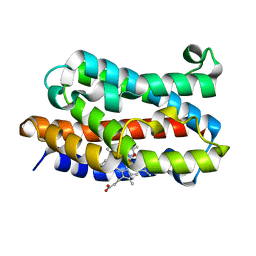 | | Crystal Structure of Rat Heme Oxygenase-1 in complex with Heme bound to Carbon Monoxide | | Descriptor: | CARBON MONOXIDE, HEME OXYGENASE-1, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Sugishima, M, Sakamoto, H, Omata, Y, Hayashi, S, Noguchi, M, Fukuyama, K. | | Deposit date: | 2002-06-10 | | Release date: | 2003-09-02 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structures of Ferrous and CO-, CN(-)-, and NO-Bound Forms of Rat Heme Oxygenase-1 (HO-1) in Complex with Heme: Structural Implications for Discrimination between CO and O(2) in HO-1.
Biochemistry, 42, 2003
|
|
1IX5
 
 | | Solution structure of the Methanococcus thermolithotrophicus FKBP | | Descriptor: | FKBP | | Authors: | Suzuki, R, Nagata, K, Kawakami, M, Nemoto, N, Furutani, M, Adachi, K, Maruyama, T, Tanokura, M. | | Deposit date: | 2002-06-12 | | Release date: | 2003-06-10 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional Solution Structure of an Archaeal FKBP with a Dual Function of Peptidyl Prolyl cis-trans Isomerase and Chaperone-like Activities
J.MOL.BIOL., 328, 2003
|
|
1IX6
 
 | | Aspartate Aminotransferase Active Site Mutant V39F | | Descriptor: | Aspartate Aminotransferase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Hayashi, H, Mizuguchi, H, Miyahara, I, Nakajima, Y, Hirotsu, K, Kagamiyama, H. | | Deposit date: | 2002-06-14 | | Release date: | 2002-07-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Conformational change in aspartate aminotransferase on substrate binding induces strain in the catalytic group and enhances catalysis
J.BIOL.CHEM., 278, 2003
|
|
1IX7
 
 | | Aspartate Aminotransferase Active Site Mutant V39F maleate complex | | Descriptor: | Aspartate Aminotransferase, MALEIC ACID, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Hayashi, H, Mizuguchi, H, Miyahara, I, Nakajima, Y, Hirotsu, K, Kagamiyama, H. | | Deposit date: | 2002-06-14 | | Release date: | 2002-07-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Conformational change in aspartate aminotransferase on substrate binding induces strain in the catalytic group and enhances catalysis
J.BIOL.CHEM., 278, 2003
|
|
1IX8
 
 | | Aspartate Aminotransferase Active Site Mutant V39F/N194A | | Descriptor: | Aspartate Aminotransferase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Hayashi, H, Mizuguchi, H, Miyahara, I, Nakajima, Y, Hirotsu, K, Kagamiyama, H. | | Deposit date: | 2002-06-14 | | Release date: | 2002-07-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Conformational change in aspartate aminotransferase on substrate binding induces strain in the catalytic group and enhances catalysis
J.BIOL.CHEM., 278, 2003
|
|
1IX9
 
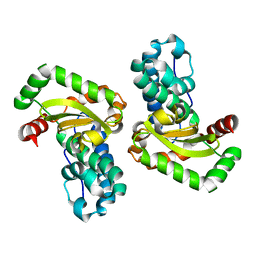 | | Crystal Structure of the E. coli Manganase(III) superoxide dismutase mutant Y174F at 0.90 angstroms resolution. | | Descriptor: | MANGANESE (II) ION, Superoxide Dismutase | | Authors: | Anderson, B.F, Edwards, R.A, Whittaker, M.M, Whittaker, J.W, Baker, E.N, Jameson, G.B. | | Deposit date: | 2002-06-17 | | Release date: | 2002-12-17 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (0.9 Å) | | Cite: | Structures at 0.90 A resolution of the oxidised and reduced forms of the Y174F mutant of the manganese superoxide dismutase from Escherichia coli
To be Published
|
|
1IXA
 
 | | THE THREE-DIMENSIONAL STRUCTURE OF THE FIRST EGF-LIKE MODULE OF HUMAN FACTOR IX: COMPARISON WITH EGF AND TGF-A | | Descriptor: | EGF-LIKE MODULE OF HUMAN FACTOR IX | | Authors: | Baron, M, Norman, D.G, Harvey, T.S, Hanford, P.A, Mayhew, M, Tse, A.G.D, Brownlee, G.G, Campbell, I.D.C. | | Deposit date: | 1991-11-14 | | Release date: | 1993-10-31 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | The three-dimensional structure of the first EGF-like module of human factor IX: comparison with EGF and TGF-alpha.
Protein Sci., 1, 1992
|
|
1IXB
 
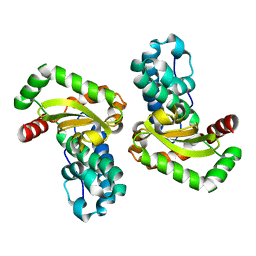 | | CRYSTAL STRUCTURE OF THE E. COLI MANGANESE(II) SUPEROXIDE DISMUTASE MUTANT Y174F AT 0.90 ANGSTROMS RESOLUTION. | | Descriptor: | MANGANESE ION, 1 HYDROXYL COORDINATED, SUPEROXIDE DISMUTASE | | Authors: | Anderson, B.F, Edwards, R.A, Whittaker, M.M, Whittaker, J.W, Baker, E.N, Jameson, G.B. | | Deposit date: | 2002-06-18 | | Release date: | 2002-12-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (0.9 Å) | | Cite: | Structures at 0.90 A resolution of the oxidised and reduced forms of the Y174F mutant of the manganese superoxide dismutase from Escherichia coli
To be Published
|
|
1IXC
 
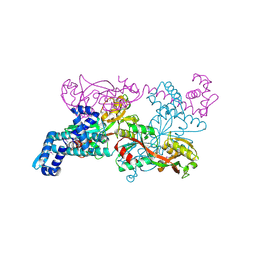 | | Crystal structure of CbnR, a LysR family transcriptional regulator | | Descriptor: | LysR-type regulatory protein | | Authors: | Muraoka, S, Okumura, R, Ogawa, N, Miyashita, K, Senda, T. | | Deposit date: | 2002-06-18 | | Release date: | 2003-06-18 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of a Full-length LysR-type Transcriptional Regulator, CbnR: Unusual Combination of Two Subunit Forms and Molecular Bases for Causing and Changing DNA Bend
J.Mol.Biol., 328, 2003
|
|
1IXD
 
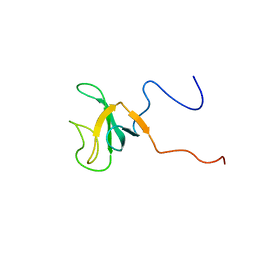 | | Solution structure of the CAP-GLY domain from human cylindromatosis tomour-suppressor CYLD | | Descriptor: | Cylindromatosis tumour-suppressor CYLD | | Authors: | Saito, K, Koshiba, S, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2002-06-19 | | Release date: | 2002-12-19 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | The CAP-Gly domain of CYLD associates with the proline-rich sequence in NEMO/IKKgamma
STRUCTURE, 12, 2004
|
|
1IXE
 
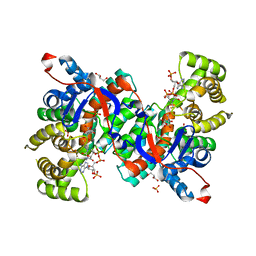 | | Crystal structure of citrate synthase from Thermus thermophilus HB8 | | Descriptor: | CITRIC ACID, COENZYME A, GLYCEROL, ... | | Authors: | Murakami, M, Kanamori, E, Kawaguchi, S, Kuramitsu, S, Kouyama, T, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2002-06-20 | | Release date: | 2003-07-29 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural comparison between the open and closed forms of citrate synthase from Thermus thermophilus HB8.
Biophys Physicobio., 12, 2015
|
|
1IXF
 
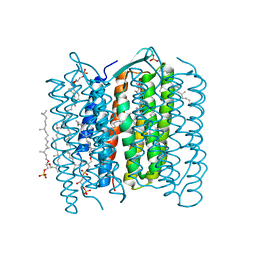 | | Crystal Structure of the K intermediate of bacteriorhodopsin | | Descriptor: | 2,3-DI-O-PHYTANLY-3-SN-GLYCERO-1-PHOSPHORYL-3'-SN-GLYCEROL-1'-PHOSPHATE, 2,3-DI-PHYTANYL-GLYCEROL, RETINAL, ... | | Authors: | Matsui, Y, Sakai, K, Murakami, M, Shiro, Y, Adachi, S, Okumura, H, Kouyama, T, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2002-06-20 | | Release date: | 2002-12-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Specific Damage Induced by X-ray Radiation and Structural Changes in the Primary Photoreaction of Bacteriorhodopsin
J.MOL.BIOL., 324, 2002
|
|
1IXG
 
 | |
1IXH
 
 | |
1IXI
 
 | |
1IXJ
 
 | | Crystal Structure of d(GCGAAAGCT) Containing Parallel-stranded Duplex with Homo Base Pairs and Anti-Parallel Duplex with Watson-Crick Base pairs | | Descriptor: | 5'-D(*GP*CP*GP*AP*AP*AP*GP*CP*T)-3', COBALT HEXAMMINE(III), MAGNESIUM ION | | Authors: | Sunami, T, Kondo, J, Kobuna, T, Hirao, I, Watanabe, K, Miura, K, Takenaka, A. | | Deposit date: | 2002-06-22 | | Release date: | 2002-12-11 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of d(GCGAAAGCT) Containing a Parallel-stranded Duplex with Homo Base Pairs and an Anti-Parallel Duplex with Watson-Crick Base pairs
Nucleic Acids Res., 30, 2002
|
|
1IXK
 
 | | Crystal Structure Analysis of Methyltransferase Homolog Protein from Pyrococcus Horikoshii | | Descriptor: | Methyltransferase | | Authors: | Ishikawa, I, Sakai, N, Yao, M, Watanabe, N, Tamura, T, Tanaka, I. | | Deposit date: | 2002-06-25 | | Release date: | 2003-09-09 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of human p120 homologue protein PH1374 from Pyrococcus horikoshii
PROTEINS: STRUCT.,FUNCT.,GENET., 54, 2004
|
|
1IXL
 
 | | Crystal structure of uncharacterized protein PH1136 from Pyrococcus horikoshii | | Descriptor: | hypothetical protein PH1136 | | Authors: | Tajika, Y, Sakai, N, Tanaka, Y, Yao, M, Watanabe, N, Tanaka, I. | | Deposit date: | 2002-06-27 | | Release date: | 2003-09-09 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Crystal structure of conserved protein PH1136 from Pyrococcus horikoshii.
Proteins, 55, 2004
|
|
