1IDC
 
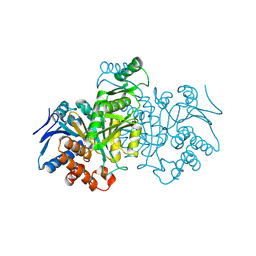 | | ISOCITRATE DEHYDROGENASE FROM E.COLI (MUTANT K230M), STEADY-STATE INTERMEDIATE COMPLEX DETERMINED BY LAUE CRYSTALLOGRAPHY | | Descriptor: | 2-OXALOSUCCINIC ACID, ISOCITRATE DEHYDROGENASE, MAGNESIUM ION | | Authors: | Bolduc, J.M, Dyer, D.H, Scott, W.G, Singer, P, Sweet, R.M, Koshland Junior, D.E, Stoddard, B.L. | | Deposit date: | 1995-01-18 | | Release date: | 1996-03-08 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Mutagenesis and Laue structures of enzyme intermediates: isocitrate dehydrogenase.
Science, 268, 1995
|
|
1IDD
 
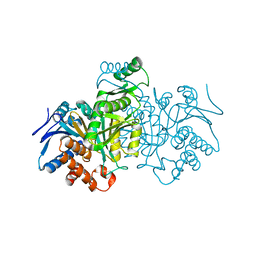 | | ISOCITRATE DEHYDROGENASE Y160F MUTANT APO ENZYME | | Descriptor: | ISOCITRATE DEHYDROGENASE | | Authors: | Lee, M.E, Dyer, D.H, Klein, O.D, Bolduc, J.M, Stoddard, B.L, Koshland Junior, D.E. | | Deposit date: | 1995-01-18 | | Release date: | 1996-03-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Mutagenesis and Laue structures of enzyme intermediates: isocitrate dehydrogenase.
Science, 268, 1995
|
|
1IDE
 
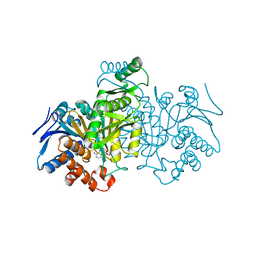 | | ISOCITRATE DEHYDROGENASE Y160F MUTANT STEADY-STATE INTERMEDIATE COMPLEX (LAUE DETERMINATION) | | Descriptor: | ISOCITRATE DEHYDROGENASE, ISOCITRIC ACID, MAGNESIUM ION, ... | | Authors: | Bolduc, J.M, Dyer, D.H, Scott, W.G, Singer, P, Sweet, R.M, Koshland Junior, D.E, Stoddard, B.L. | | Deposit date: | 1995-01-18 | | Release date: | 1996-03-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Mutagenesis and Laue structures of enzyme intermediates: isocitrate dehydrogenase.
Science, 268, 1995
|
|
1IDF
 
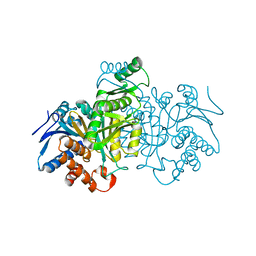 | | ISOCITRATE DEHYDROGENASE K230M MUTANT APO ENZYME | | Descriptor: | ISOCITRATE DEHYDROGENASE | | Authors: | Bolduc, J.M, Dyer, D.H, Scott, W.G, Singer, P, Sweet, R.M, Koshland Junior, D.E, Stoddard, B.L. | | Deposit date: | 1995-01-18 | | Release date: | 1996-03-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Mutagenesis and Laue structures of enzyme intermediates: isocitrate dehydrogenase.
Science, 268, 1995
|
|
1IDG
 
 | | THE NMR SOLUTION STRUCTURE OF THE COMPLEX FORMED BETWEEN ALPHA-BUNGAROTOXIN AND AN 18MER COGNATE PEPTIDE | | Descriptor: | ACETYLCHOLINE RECEPTOR PROTEIN, ALPHA CHAIN, ALPHA-BUNGAROTOXIN | | Authors: | Zeng, H, Moise, L, Grant, M.A, Hawrot, E. | | Deposit date: | 2001-04-04 | | Release date: | 2001-04-25 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | The solution structure of the complex formed between alpha-bungarotoxin and an 18-mer cognate peptide derived from the alpha 1 subunit of the nicotinic acetylcholine receptor from Torpedo californica.
J.Biol.Chem., 276, 2001
|
|
1IDH
 
 | | THE NMR SOLUTION STRUCTURE OF THE COMPLEX FORMED BETWEEN ALPHA-BUNGAROTOXIN AND AN 18MER COGNATE PEPTIDE | | Descriptor: | ACETYLCHOLINE RECEPTOR PROTEIN, ALPHA CHAIN, ALPHA-BUNGAROTOXIN | | Authors: | Zeng, H, Moise, L, Grant, M.A, Hawrot, E. | | Deposit date: | 2001-04-04 | | Release date: | 2001-04-25 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | The solution structure of the complex formed between alpha-bungarotoxin and an 18-mer cognate peptide derived from the alpha 1 subunit of the nicotinic acetylcholine receptor from Torpedo californica.
J.Biol.Chem., 276, 2001
|
|
1IDI
 
 | | THE NMR SOLUTION STRUCTURE OF ALPHA-BUNGAROTOXIN | | Descriptor: | ALPHA-BUNGAROTOXIN | | Authors: | Zeng, H, Moise, L, Grant, M.A, Hawrot, E. | | Deposit date: | 2001-04-04 | | Release date: | 2001-04-25 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | The solution structure of the complex formed between alpha-bungarotoxin and an 18-mer cognate peptide derived from the alpha 1 subunit of the nicotinic acetylcholine receptor from Torpedo californica.
J.Biol.Chem., 276, 2001
|
|
1IDJ
 
 | | PECTIN LYASE A | | Descriptor: | PECTIN LYASE A | | Authors: | Mayans, O, Scott, M, Connerton, I, Gravesen, T, Benen, J, Visser, J, Pickersgill, R, Jenkins, J. | | Deposit date: | 1996-10-04 | | Release date: | 1997-10-15 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Two crystal structures of pectin lyase A from Aspergillus reveal a pH driven conformational change and striking divergence in the substrate-binding clefts of pectin and pectate lyases.
Structure, 5, 1997
|
|
1IDK
 
 | | PECTIN LYASE A | | Descriptor: | PECTIN LYASE A | | Authors: | Mayans, O, Scott, M, Connerton, I, Gravesen, T, Benen, J, Visser, J, Pickersgill, R, Jenkins, J. | | Deposit date: | 1996-10-04 | | Release date: | 1997-10-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Two crystal structures of pectin lyase A from Aspergillus reveal a pH driven conformational change and striking divergence in the substrate-binding clefts of pectin and pectate lyases.
Structure, 5, 1997
|
|
1IDL
 
 | | THE NMR SOLUTION STRUCTURE OF ALPHA-BUNGAROTOXIN | | Descriptor: | ALPHA-BUNGAROTOXIN | | Authors: | Zeng, H, Moise, L, Grant, M.A, Hawrot, E. | | Deposit date: | 2001-04-04 | | Release date: | 2001-04-25 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | The solution structure of the complex formed between alpha-bungarotoxin and an 18-mer cognate peptide derived from the alpha 1 subunit of the nicotinic acetylcholine receptor from Torpedo californica.
J.Biol.Chem., 276, 2001
|
|
1IDM
 
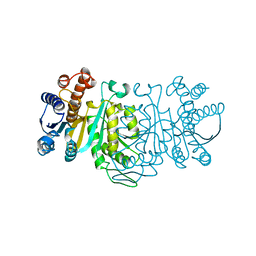 | | 3-ISOPROPYLMALATE DEHYDROGENASE, LOOP-DELETED CHIMERA | | Descriptor: | 3-ISOPROPYLMALATE DEHYDROGENASE | | Authors: | Sakurai, M, Ohzeki, M, Moriyama, H, Sato, M, Tanaka, N. | | Deposit date: | 1995-05-19 | | Release date: | 1995-09-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of a loop-deleted variant of 3-isopropylmalate dehydrogenase from Thermus thermophilus: an internal reprieve tolerance mechanism.
Acta Crystallogr.,Sect.D, 52, 1996
|
|
1IDN
 
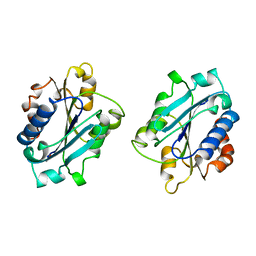 | | MAC-1 I DOMAIN METAL FREE | | Descriptor: | CD11B | | Authors: | Baldwin, E.T. | | Deposit date: | 1998-06-10 | | Release date: | 1998-11-04 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Cation binding to the integrin CD11b I domain and activation model assessment
Structure, 6, 1998
|
|
1IDO
 
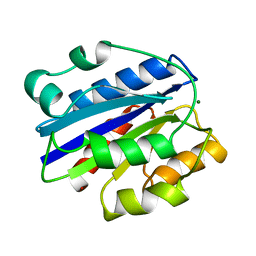 | | I-DOMAIN FROM INTEGRIN CR3, MG2+ BOUND | | Descriptor: | INTEGRIN, MAGNESIUM ION | | Authors: | Lee, J.-O, Liddington, R. | | Deposit date: | 1996-03-12 | | Release date: | 1996-08-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of the A domain from the alpha subunit of integrin CR3 (CD11b/CD18).
Cell(Cambridge,Mass.), 80, 1995
|
|
1IDP
 
 | |
1IDQ
 
 | |
1IDR
 
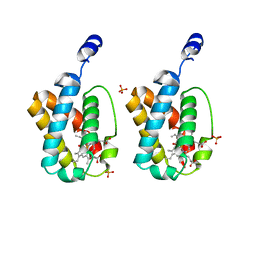 | | CRYSTAL STRUCTURE OF THE TRUNCATED-HEMOGLOBIN-N FROM MYCOBACTERIUM TUBERCULOSIS | | Descriptor: | HEMOGLOBIN HBN, OXYGEN MOLECULE, PHOSPHATE ION, ... | | Authors: | Milani, M, Pesce, A, Ascenzi, P, Guertin, M, Bolognesi, M. | | Deposit date: | 2001-04-05 | | Release date: | 2001-08-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Mycobacterium tuberculosis hemoglobin N displays a protein tunnel suited for O2 diffusion to the heme.
EMBO J., 20, 2001
|
|
1IDS
 
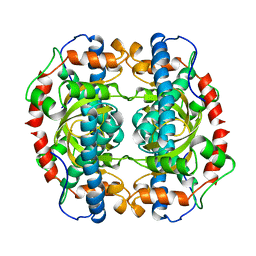 | | X-RAY STRUCTURE ANALYSIS OF THE IRON-DEPENDENT SUPEROXIDE DISMUTASE FROM MYCOBACTERIUM TUBERCULOSIS AT 2.0 ANGSTROMS RESOLUTIONS REVEALS NOVEL DIMER-DIMER INTERACTIONS | | Descriptor: | FE (III) ION, IRON SUPEROXIDE DISMUTASE | | Authors: | Cooper, J.B, Mcintyre, K, Wood, S.P, Zhang, Y, Young, D. | | Deposit date: | 1994-09-29 | | Release date: | 1994-12-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-ray structure analysis of the iron-dependent superoxide dismutase from Mycobacterium tuberculosis at 2.0 Angstroms resolution reveals novel dimer-dimer interactions.
J.Mol.Biol., 246, 1995
|
|
1IDT
 
 | | STRUCTURAL STUDIES ON A PRODRUG-ACTIVATING SYSTEM-CB1954 AND FMN-DEPENDENT NITROREDUCTASE | | Descriptor: | 5-(AZIRIDIN-1-YL)-2,4-DINITROBENZAMIDE, FLAVIN MONONUCLEOTIDE, MINOR FMN-DEPENDENT NITROREDUCTASE | | Authors: | Johansson, E, Parkinson, G.N, Denny, W.A, Neidle, S. | | Deposit date: | 2001-04-05 | | Release date: | 2003-09-16 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Studies on the Nitroreductase Prodrug-Activating System. Crystal Structures of Complexes with the Inhibitor Dicoumarol and Dinitrobenzamide Prodrugs and of the Enzyme Active Form.
J.Med.Chem., 46, 2003
|
|
1IDU
 
 | |
1IDV
 
 | |
1IDW
 
 | | STRUCTURE OF THE HYBRID RNA/DNA R-GCUUCGGC-D[CL]U IN PRESENCE OF RH(NH3)6+++ | | Descriptor: | 5'-R(*GP*CP*UP*UP*CP*GP*GP*C)-D(P*(UCL))-3', CHLORIDE ION, RHODIUM HEXAMINE ION | | Authors: | Cruse, W, Saludjian, P, Neuman, A, Prange, T. | | Deposit date: | 2001-04-05 | | Release date: | 2001-04-12 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Destabilizing effect of a fluorouracil extra base in a hybrid RNA duplex compared with bromo and chloro analogues
Acta Crystallogr.,Sect.D, 57, 2001
|
|
1IDX
 
 | |
1IDY
 
 | |
1IDZ
 
 | |
1IE0
 
 | | CRYSTAL STRUCTURE OF LUXS | | Descriptor: | AUTOINDUCER-2 PRODUCTION PROTEIN LUXS, GLYCEROL, ZINC ION | | Authors: | Hilgers, M.T, Ludwig, M.L. | | Deposit date: | 2001-04-05 | | Release date: | 2001-10-03 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of the quorum-sensing protein LuxS reveals a catalytic metal site.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
