1HV6
 
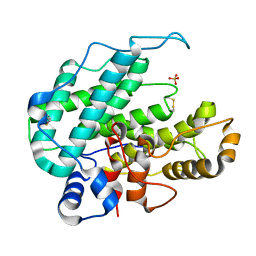 | | CRYSTAL STRUCTURE OF ALGINATE LYASE A1-III COMPLEXED WITH TRISACCHARIDE PRODUCT. | | 分子名称: | 4-deoxy-alpha-L-erythro-hex-4-enopyranuronic acid-(1-4)-alpha-D-mannopyranuronic acid-(1-4)-alpha-D-glucopyranuronic acid, ALGINATE LYASE, SULFATE ION | | 著者 | Yoon, H.-J, Hashimoto, W, Miyake, O, Murata, K, Mikami, B. | | 登録日 | 2001-01-08 | | 公開日 | 2001-05-02 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Crystal structure of alginate lyase A1-III complexed with trisaccharide product at 2.0 A resolution.
J.Mol.Biol., 307, 2001
|
|
7SA8
 
 | |
1QAZ
 
 | |
8BDQ
 
 | |
4E1Y
 
 | | Alginate lyase A1-III H192A apo form | | 分子名称: | Alginate lyase | | 著者 | Mikami, B, Ban, M, Suzuki, S, Yoon, H.-J, Miyake, O, Yamasaki, M, Ogura, K, Maruyama, Y, Hashimoto, W, Murata, K. | | 登録日 | 2012-03-07 | | 公開日 | 2012-04-11 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Induced-fit motion of a lid loop involved in catalysis in alginate lyase A1-III
Acta Crystallogr.,Sect.D, 68, 2012
|
|
4F13
 
 | | Alginate lyase A1-III Y246F complexed with tetrasaccharide | | 分子名称: | 4-deoxy-alpha-L-erythro-hex-4-enopyranuronic acid-(1-4)-beta-D-mannopyranuronic acid-(1-4)-beta-D-mannopyranuronic acid-(1-4)-beta-D-mannopyranuronic acid, Alginate lyase, beta-D-mannopyranuronic acid-(1-4)-beta-D-mannopyranuronic acid | | 著者 | Mikami, B, Ban, M, Suzuki, S, Yoon, H.-J, Miyake, O, Yamasaki, M, Ogura, K, Maruyama, Y, Hashimoto, W, Murata, K. | | 登録日 | 2012-05-06 | | 公開日 | 2012-06-27 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.208 Å) | | 主引用文献 | Induced-fit motion of a lid loop involved in catalysis in alginate lyase A1-III
Acta Crystallogr.,Sect.D, 68, 2012
|
|
4F10
 
 | | Alginate lyase A1-III H192A complexed with tetrasaccharide | | 分子名称: | 4-deoxy-alpha-L-erythro-hex-4-enopyranuronic acid-(1-4)-alpha-L-gulopyranuronic acid-(1-4)-beta-D-mannopyranuronic acid-(1-4)-beta-D-mannopyranuronic acid, Alginate lyase, alpha-L-gulopyranuronic acid-(1-4)-beta-D-mannopyranuronic acid | | 著者 | Mikami, B, Ban, M, Suzuki, S, Yoon, H.-J, Miyake, O, Yamasaki, M, Ogura, K, Maruyama, Y, Hashimoto, W, Murata, K. | | 登録日 | 2012-05-05 | | 公開日 | 2012-06-27 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Induced-fit motion of a lid loop involved in catalysis in alginate lyase A1-III
Acta Crystallogr.,Sect.D, 68, 2012
|
|
4OZV
 
 | | Crystal Structure of the periplasmic alginate lyase AlgL | | 分子名称: | Alginate lyase, beta-D-mannopyranuronic acid | | 著者 | Howell, P.L, Wolfram, F, Robinson, H, Arora, K. | | 登録日 | 2014-02-19 | | 公開日 | 2015-03-04 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.642 Å) | | 主引用文献 | The Pseudomonas aeruginosa homeostasis enzyme AlgL clears the periplasmic space of accumulated alginate during polymer biosynthesis.
J.Biol.Chem., 298, 2022
|
|
4OZW
 
 | |
7FHZ
 
 | |
7FI1
 
 | |
7FI2
 
 | |
7FI0
 
 | | Crystal structure of Multi-functional Polysaccharide lyase Smlt1473 (WT) from Stenotrophomonas maltophilia (strain K279a) in ManA bound form at pH-5.0 | | 分子名称: | DI(HYDROXYETHYL)ETHER, Polysaccharide lyase, SULFATE ION, ... | | 著者 | Pandey, S, Berger, B.W, Acharya, R. | | 登録日 | 2021-07-30 | | 公開日 | 2021-10-27 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.31 Å) | | 主引用文献 | Structural insights into the mechanism of pH-selective substrate specificity of the polysaccharide lyase Smlt1473.
J.Biol.Chem., 297, 2021
|
|
7FHY
 
 | |
7FHX
 
 | |
7FHU
 
 | |
7FHV
 
 | |
7FHW
 
 | |
7XTF
 
 | |
7XTE
 
 | |
7WXM
 
 | |
7WXN
 
 | |
7WXR
 
 | |
7WXP
 
 | |
7WXL
 
 | |
