1F17
 
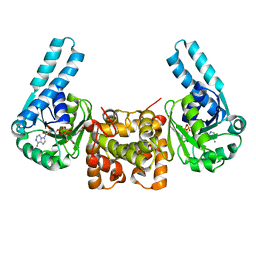 | | L-3-HYDROXYACYL-COA DEHYDROGENASE COMPLEXED WITH NADH | | 分子名称: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, L-3-HYDROXYACYL-COA DEHYDROGENASE | | 著者 | Barycki, J.J, O'Brien, L.K, Strauss, A.W, Banaszak, L.J. | | 登録日 | 2000-05-18 | | 公開日 | 2000-09-27 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Sequestration of the active site by interdomain shifting. Crystallographic and spectroscopic evidence for distinct conformations of L-3-hydroxyacyl-CoA dehydrogenase.
J.Biol.Chem., 275, 2000
|
|
1F12
 
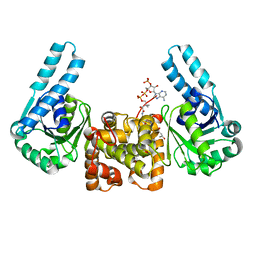 | | L-3-HYDROXYACYL-COA DEHYDROGENASE COMPLEXED WITH 3-HYDROXYBUTYRYL-COA | | 分子名称: | 3-HYDROXYBUTANOYL-COENZYME A, L-3-HYDROXYACYL-COA DEHYDROGENASE | | 著者 | Barycki, J.J, O'Brien, L.K, Strauss, A.W, Banaszak, L.J. | | 登録日 | 2000-05-18 | | 公開日 | 2000-09-27 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Sequestration of the active site by interdomain shifting. Crystallographic and spectroscopic evidence for distinct conformations of L-3-hydroxyacyl-CoA dehydrogenase.
J.Biol.Chem., 275, 2000
|
|
3WAJ
 
 | |
3WAK
 
 | |
1OR0
 
 | | Crystal Structures of Glutaryl 7-Aminocephalosporanic Acid Acylase: Insight into Autoproteolytic Activation | | 分子名称: | 1,2-ETHANEDIOL, Glutaryl 7-Aminocephalosporanic Acid Acylase, glutaryl acylase | | 著者 | Kim, J.K, Yang, I.S, Rhee, S, Dauter, Z, Lee, Y.S, Park, S.S, Kim, K.H. | | 登録日 | 2003-03-11 | | 公開日 | 2004-03-11 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Crystal Structures of Glutaryl 7-Aminocephalosporanic Acid Acylase: Insight into Autoproteolytic Activation
Biochemistry, 42, 2003
|
|
1ANG
 
 | | CRYSTAL STRUCTURE OF HUMAN ANGIOGENIN REVEALS THE STRUCTURAL BASIS FOR ITS FUNCTIONAL DIVERGENCE FROM RIBONUCLEASE | | 分子名称: | ANGIOGENIN | | 著者 | Acharya, K.R, Allen, S, Shapiro, R, Riordan, J.F, Vallee, B.L. | | 登録日 | 1994-01-18 | | 公開日 | 1995-04-20 | | 最終更新日 | 2024-10-09 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Crystal structure of human angiogenin reveals the structural basis for its functional divergence from ribonuclease.
Proc.Natl.Acad.Sci.USA, 91, 1994
|
|
1F0Y
 
 | | L-3-HYDROXYACYL-COA DEHYDROGENASE COMPLEXED WITH ACETOACETYL-COA AND NAD+ | | 分子名称: | ACETOACETYL-COENZYME A, L-3-HYDROXYACYL-COA DEHYDROGENASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | 著者 | Barycki, J.J, O'Brien, L.K, Strauss, A.W, Banaszak, L.J. | | 登録日 | 2000-05-17 | | 公開日 | 2000-09-01 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Sequestration of the active site by interdomain shifting. Crystallographic and spectroscopic evidence for distinct conformations of L-3-hydroxyacyl-CoA dehydrogenase.
J.Biol.Chem., 275, 2000
|
|
1BZO
 
 | | THREE-DIMENSIONAL STRUCTURE OF PROKARYOTIC CU,ZN SUPEROXIDE DISMUTASE FROM P.LEIOGNATHI, SOLVED BY X-RAY CRYSTALLOGRAPHY. | | 分子名称: | COPPER (II) ION, PROTEIN (SUPEROXIDE DISMUTASE), URANYL (VI) ION, ... | | 著者 | Bordo, D, Matak, D, Djinovic-Carugo, K, Rosano, C, Pesce, A, Bolognesi, M, Stroppolo, M.E, Falconi, M, Battistoni, A, Desideri, A. | | 登録日 | 1998-11-02 | | 公開日 | 1999-04-09 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Evolutionary constraints for dimer formation in prokaryotic Cu,Zn superoxide dismutase.
J.Mol.Biol., 285, 1999
|
|
4OY2
 
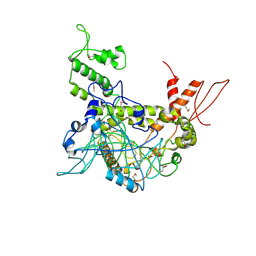 | | Crystal structure of TAF1-TAF7, a TFIID subcomplex | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Transcription initiation factor TFIID subunit 1, Transcription initiation factor TFIID subunit 7, ... | | 著者 | Bhattacharya, S, Lou, X, Rajashankar, K, Jacobson, R.H, Webb, P. | | 登録日 | 2014-02-10 | | 公開日 | 2014-06-25 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Structural and functional insight into TAF1-TAF7, a subcomplex of transcription factor II D.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
6LVB
 
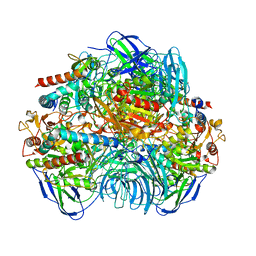 | | Structure of Dimethylformamidase, tetramer | | 分子名称: | FE (III) ION, N,N-dimethylformamidase large subunit, N,N-dimethylformamidase small subunit | | 著者 | Arya, C.A, Yadav, S, Fine, J, Casanal, A, Chopra, G, Ramanathan, G, Subramanian, R, Vinothkumar, K.R. | | 登録日 | 2020-02-02 | | 公開日 | 2020-06-03 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (2.8 Å) | | 主引用文献 | A 2-Tyr-1-carboxylate Mononuclear Iron Center Forms the Active Site of a Paracoccus Dimethylformamidase.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
6LVE
 
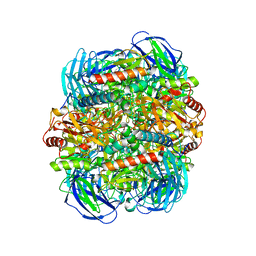 | | Structure of Dimethylformamidase, tetramer, E521A mutant | | 分子名称: | N,N-dimethylformamidase large subunit, N,N-dimethylformamidase small subunit | | 著者 | Arya, C.A, Yadav, S, Fine, J, Casanal, A, Chopra, G, Ramanathan, G, Subramanian, R, Vinothkumar, K.R. | | 登録日 | 2020-02-02 | | 公開日 | 2020-06-03 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | A 2-Tyr-1-carboxylate Mononuclear Iron Center Forms the Active Site of a Paracoccus Dimethylformamidase.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
6LVC
 
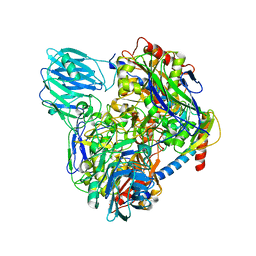 | | Structure of Dimethylformamidase, dimer | | 分子名称: | FE (III) ION, N,N-dimethylformamidase large subunit, N,N-dimethylformamidase small subunit | | 著者 | Arya, C.A, Yadav, S, Fine, J, Casanal, A, Chopra, G, Ramanathan, G, Subramanian, R, Vinothkumar, K.R. | | 登録日 | 2020-02-02 | | 公開日 | 2020-06-03 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | A 2-Tyr-1-carboxylate Mononuclear Iron Center Forms the Active Site of a Paracoccus Dimethylformamidase.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
1A9V
 
 | |
6LVV
 
 | | N, N-dimethylformamidase | | 分子名称: | 1,2-ETHANEDIOL, FE (III) ION, N,N-dimethylformamidase large subunit, ... | | 著者 | Arya, C.K, Ramaswamy, S, Kutti, R.V, Gurunath, R. | | 登録日 | 2020-02-05 | | 公開日 | 2020-08-12 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | A 2-Tyr-1-carboxylate Mononuclear Iron Center Forms the Active Site of a Paracoccus Dimethylformamidase.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
7SOM
 
 | | Ciliary C2 central pair apparatus isolated from Chlamydomonas reinhardtii | | 分子名称: | Cilia- and flagella-associated protein 20, FAP147, FAP178, ... | | 著者 | Gui, M, Wang, X, Dutcher, S.K, Brown, A, Zhang, R. | | 登録日 | 2021-11-01 | | 公開日 | 2022-04-13 | | 最終更新日 | 2024-06-05 | | 実験手法 | ELECTRON MICROSCOPY (3.7 Å) | | 主引用文献 | Ciliary central apparatus structure reveals mechanisms of microtubule patterning.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7SQC
 
 | | Ciliary C1 central pair apparatus isolated from Chlamydomonas reinhardtii | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, CPC1, Calmodulin, ... | | 著者 | Gui, M, Wang, X, Dutcher, S.K, Brown, A, Zhang, R. | | 登録日 | 2021-11-05 | | 公開日 | 2022-04-13 | | 最終更新日 | 2024-06-05 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | Ciliary central apparatus structure reveals mechanisms of microtubule patterning.
Nat.Struct.Mol.Biol., 29, 2022
|
|
6LVD
 
 | | Structure of Dimethylformamidase, tetramer, Y440A mutant | | 分子名称: | N,N-dimethylformamidase large subunit, N,N-dimethylformamidase small subunit | | 著者 | Arya, C.A, Yadav, S, Fine, J, Casanal, A, Chopra, G, Ramanathan, G, Subramanian, R, Vinothkumar, K.R. | | 登録日 | 2020-02-02 | | 公開日 | 2020-06-03 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | A 2-Tyr-1-carboxylate Mononuclear Iron Center Forms the Active Site of a Paracoccus Dimethylformamidase.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
1SMD
 
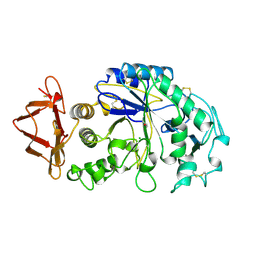 | | HUMAN SALIVARY AMYLASE | | 分子名称: | AMYLASE, CALCIUM ION, CHLORIDE ION | | 著者 | Ramasubbu, N. | | 登録日 | 1996-01-24 | | 公開日 | 1996-07-11 | | 最終更新日 | 2019-12-25 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Structure of human salivary alpha-amylase at 1.6 A resolution: implications for its role in the oral cavity.
Acta Crystallogr.,Sect.D, 52, 1996
|
|
4REK
 
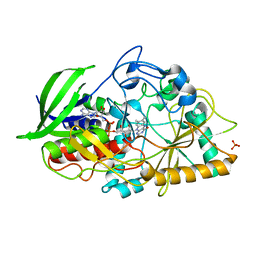 | | Crystal structure and charge density studies of cholesterol oxidase from Brevibacterium sterolicum at 0.74 ultra-high resolution | | 分子名称: | Cholesterol oxidase, FLAVIN-ADENINE DINUCLEOTIDE, PHOSPHATE ION | | 著者 | Zarychta, B, Lyubimov, A, Ahmed, M, Munshi, P, Guillot, B, Vrielink, A. | | 登録日 | 2014-09-23 | | 公開日 | 2015-04-15 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (0.74 Å) | | 主引用文献 | Cholesterol oxidase: ultrahigh-resolution crystal structure and multipolar atom model-based analysis.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
1ALC
 
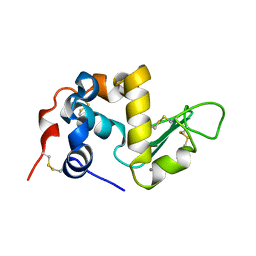 | |
7S5J
 
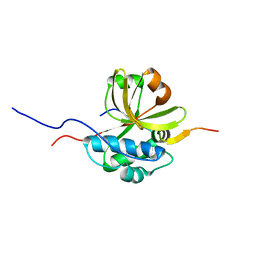 | |
1CNY
 
 | |
1CNX
 
 | |
1BBT
 
 | | METHODS USED IN THE STRUCTURE DETERMINATION OF FOOT AND MOUTH DISEASE VIRUS | | 分子名称: | FOOT-AND-MOUTH DISEASE VIRUS (SUBUNIT VP1), FOOT-AND-MOUTH DISEASE VIRUS (SUBUNIT VP2), FOOT-AND-MOUTH DISEASE VIRUS (SUBUNIT VP3), ... | | 著者 | Acharya, K.R, Fry, E.E, Logan, D.T, Stuart, D.I. | | 登録日 | 1992-05-18 | | 公開日 | 1994-01-31 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Methods used in the structure determination of foot-and-mouth disease virus.
Acta Crystallogr.,Sect.A, 49, 1993
|
|
3MPJ
 
 | | Structure of the glutaryl-coenzyme A dehydrogenase | | 分子名称: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, Glutaryl-CoA dehydrogenase, ... | | 著者 | Wischgoll, S, Warkentin, E, Boll, M, Ermler, U. | | 登録日 | 2010-04-27 | | 公開日 | 2010-08-18 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Structural basis for promoting and preventing decarboxylation in glutaryl-coenzyme a dehydrogenases.
Biochemistry, 49, 2010
|
|
