2VVM
 
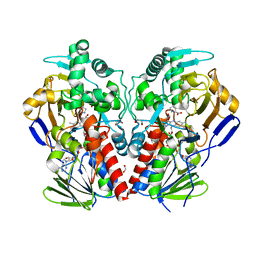 | | The structure of MAO-N-D5, a variant of monoamine oxidase from Aspergillus niger. | | 分子名称: | 1,2-ETHANEDIOL, FLAVIN-ADENINE DINUCLEOTIDE, MONOAMINE OXIDASE N, ... | | 著者 | Atkin, K.E, Hart, S, Turkenburg, J.P, Brzozowski, A.M, Grogan, G.J. | | 登録日 | 2008-06-10 | | 公開日 | 2008-11-04 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | The Structure of Monoamine Oxidase from Aspergillus Niger Provides a Molecular Context for Improvements in Activity Obtained by Directed Evolution.
J.Mol.Biol., 384, 2008
|
|
5RG8
 
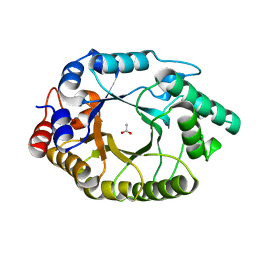 | | Crystal Structure of Kemp Eliminase HG3.17 in unbound state, 277K | | 分子名称: | ACETATE ION, Kemp Eliminase HG3 | | 著者 | Broom, A, Rakotoharisoa, R.V, Thompson, M.C, Fraser, J.S, Chica, R.A. | | 登録日 | 2020-03-19 | | 公開日 | 2020-07-22 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (1.73 Å) | | 主引用文献 | Ensemble-based enzyme design can recapitulate the effects of laboratory directed evolution in silico.
Nat Commun, 11, 2020
|
|
5RG6
 
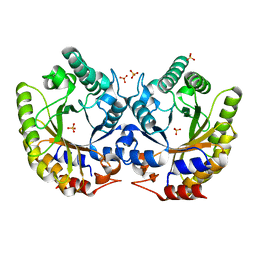 | | Crystal Structure of Kemp Eliminase HG3.7 in unbound state, 277K | | 分子名称: | Kemp Eliminase HG3, SULFATE ION | | 著者 | Broom, A, Rakotoharisoa, R.V, Thompson, M.C, Fraser, J.S, Chica, R.A. | | 登録日 | 2020-03-19 | | 公開日 | 2020-07-22 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (1.35 Å) | | 主引用文献 | Ensemble-based enzyme design can recapitulate the effects of laboratory directed evolution in silico.
Nat Commun, 11, 2020
|
|
5RG5
 
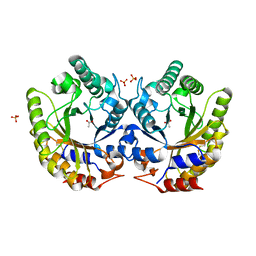 | | Crystal Structure of Kemp Eliminase HG3.3b in unbound state, 277K | | 分子名称: | ACETATE ION, Kemp Eliminase HG3, SULFATE ION | | 著者 | Broom, A, Rakotoharisoa, R.V, Thompson, M.C, Fraser, J.S, Chica, R.A. | | 登録日 | 2020-03-19 | | 公開日 | 2020-07-22 | | 最終更新日 | 2021-05-12 | | 実験手法 | X-RAY DIFFRACTION (1.62 Å) | | 主引用文献 | Ensemble-based enzyme design can recapitulate the effects of laboratory directed evolution in silico.
Nat Commun, 11, 2020
|
|
5RG7
 
 | | Crystal Structure of Kemp Eliminase HG3.14 in unbound state, 277K | | 分子名称: | Kemp Eliminase HG3, SULFATE ION | | 著者 | Broom, A, Rakotoharisoa, R.V, Thompson, M.C, Fraser, J.S, Chica, R.A. | | 登録日 | 2020-03-19 | | 公開日 | 2020-07-22 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (1.47 Å) | | 主引用文献 | Ensemble-based enzyme design can recapitulate the effects of laboratory directed evolution in silico.
Nat Commun, 11, 2020
|
|
5RGE
 
 | | Crystal Structure of Kemp Eliminase HG3.17 with bound transition state analog, 277K | | 分子名称: | 6-NITROBENZOTRIAZOLE, Kemp Eliminase HG3 | | 著者 | Broom, A, Rakotoharisoa, R.V, Thompson, M.C, Fraser, J.S, Chica, R.A. | | 登録日 | 2020-03-19 | | 公開日 | 2020-07-22 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (1.77 Å) | | 主引用文献 | Ensemble-based enzyme design can recapitulate the effects of laboratory directed evolution in silico.
Nat Commun, 11, 2020
|
|
3PO5
 
 | | Structure of a mutant of the large fragment of DNA polymerase I from Thermus Auqaticus in complex with an abasic site and ddATP | | 分子名称: | 2',3'-dideoxyadenosine triphosphate, DNA (5'-D(*GP*AP*CP*CP*AP*CP*GP*GP*CP*GP*CP*(2DA))-3'), DNA (5'-D(P*(3DR)P*TP*GP*CP*GP*CP*CP*GP*TP*GP*GP*TP*C)-3'), ... | | 著者 | Marx, A, Diederichs, K, Obeid, S. | | 登録日 | 2010-11-22 | | 公開日 | 2011-06-15 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.39 Å) | | 主引用文献 | Learning from Directed Evolution: Thermus aquaticus DNA Polymerase Mutants with Translesion Synthesis Activity.
Chembiochem, 12, 2011
|
|
7WB2
 
 | | Oxidase ChaP-D49L/Y109F mutant | | 分子名称: | ChaP, FE (III) ION | | 著者 | Zong, Y, Zheng, W, Wang, Y, Zhu, J, Tan, R. | | 登録日 | 2021-12-15 | | 公開日 | 2022-05-11 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Alteration of the Catalytic Reaction Trajectory of a Vicinal Oxygen Chelate Enzyme by Directed Evolution.
Angew.Chem.Int.Ed.Engl., 61, 2022
|
|
5RGB
 
 | | Crystal Structure of Kemp Eliminase HG3.3b with bound transition state analogue, 277K | | 分子名称: | 6-NITROBENZOTRIAZOLE, Kemp Eliminase HG3.3b, SULFATE ION | | 著者 | Broom, A, Rakotoharisoa, R.V, Thompson, M.C, Fraser, J.S, Chica, R.A. | | 登録日 | 2020-03-19 | | 公開日 | 2020-07-22 | | 最終更新日 | 2021-05-12 | | 実験手法 | X-RAY DIFFRACTION (1.42 Å) | | 主引用文献 | Ensemble-based enzyme design can recapitulate the effects of laboratory directed evolution in silico.
Nat Commun, 11, 2020
|
|
5RGD
 
 | | Crystal Structure of Kemp Eliminase HG3.14 with bound transition state analogue, 277K | | 分子名称: | 6-NITROBENZOTRIAZOLE, Kemp Eliminase HG3.14, SULFATE ION | | 著者 | Broom, A, Rakotoharisoa, R.V, Thompson, M.C, Fraser, J.S, Chica, R.A. | | 登録日 | 2020-03-19 | | 公開日 | 2020-07-22 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Ensemble-based enzyme design can recapitulate the effects of laboratory directed evolution in silico.
Nat Commun, 11, 2020
|
|
5RG4
 
 | | Crystal Structure of Kemp Eliminase HG3 in unbound state, 277K | | 分子名称: | ACETATE ION, Kemp Eliminase HG3, SULFATE ION | | 著者 | Broom, A, Rakotoharisoa, R.V, Thompson, M.C, Fraser, J.S, Chica, R.A. | | 登録日 | 2020-03-19 | | 公開日 | 2020-07-22 | | 最終更新日 | 2021-05-12 | | 実験手法 | X-RAY DIFFRACTION (1.99 Å) | | 主引用文献 | Ensemble-based enzyme design can recapitulate the effects of laboratory directed evolution in silico.
Nat Commun, 11, 2020
|
|
3PO4
 
 | | Structure of a mutant of the large fragment of DNA polymerase I from Thermus aquaticus in complex with a blunt-ended DNA and ddATP | | 分子名称: | 2',3'-dideoxyadenosine triphosphate, DNA (5'-D(*GP*AP*CP*CP*AP*CP*GP*GP*CP*GP*CP*(2DA))-3'), DNA (5'-D(*TP*GP*CP*GP*CP*CP*GP*TP*GP*GP*TP*C)-3'), ... | | 著者 | Marx, A, Diederichs, K, Obeid, S. | | 登録日 | 2010-11-22 | | 公開日 | 2011-06-15 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Learning from Directed Evolution: Thermus aquaticus DNA Polymerase Mutants with Translesion Synthesis Activity.
Chembiochem, 12, 2011
|
|
5RGC
 
 | | Crystal Structure of Kemp Eliminase HG3.7 with bound transition state analogue, 277K | | 分子名称: | 6-NITROBENZOTRIAZOLE, Kemp Eliminase HG3, SULFATE ION | | 著者 | Broom, A, Rakotoharisoa, R.V, Thompson, M.C, Fraser, J.S, Chica, R.A. | | 登録日 | 2020-03-19 | | 公開日 | 2020-07-22 | | 最終更新日 | 2021-05-12 | | 実験手法 | X-RAY DIFFRACTION (1.39 Å) | | 主引用文献 | Ensemble-based enzyme design can recapitulate the effects of laboratory directed evolution in silico.
Nat Commun, 11, 2020
|
|
5RGA
 
 | | Crystal Structure of Kemp Eliminase HG3 with bound transition state analogue, 277K | | 分子名称: | 6-NITROBENZOTRIAZOLE, Kemp Eliminase HG3, SULFATE ION | | 著者 | Broom, A, Rakotoharisoa, R.V, Thompson, M.C, Fraser, J.S, Chica, R.A. | | 登録日 | 2020-03-19 | | 公開日 | 2020-07-22 | | 最終更新日 | 2021-05-12 | | 実験手法 | X-RAY DIFFRACTION (1.86 Å) | | 主引用文献 | Ensemble-based enzyme design can recapitulate the effects of laboratory directed evolution in silico.
Nat Commun, 11, 2020
|
|
5RGF
 
 | | Crystal Structure of Kemp Eliminase HG4 with bound transition state analogue, 277K | | 分子名称: | 6-NITROBENZOTRIAZOLE, Kemp Eliminase HG3, SULFATE ION | | 著者 | Broom, A, Rakotoharisoa, R.V, Thompson, M.C, Fraser, J.S, Chica, R.A. | | 登録日 | 2020-03-19 | | 公開日 | 2020-07-22 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (1.46 Å) | | 主引用文献 | Ensemble-based enzyme design can recapitulate the effects of laboratory directed evolution in silico.
Nat Commun, 11, 2020
|
|
5RG9
 
 | | Crystal Structure of Kemp Eliminase HG4 in unbound state, 277K | | 分子名称: | ACETATE ION, Kemp Eliminase HG4, SULFATE ION | | 著者 | Broom, A, Rakotoharisoa, R.V, Thompson, M.C, Fraser, J.S, Chica, R.A. | | 登録日 | 2020-03-19 | | 公開日 | 2020-07-22 | | 最終更新日 | 2021-05-12 | | 実験手法 | X-RAY DIFFRACTION (1.47 Å) | | 主引用文献 | Ensemble-based enzyme design can recapitulate the effects of laboratory directed evolution in silico.
Nat Commun, 11, 2020
|
|
7FDY
 
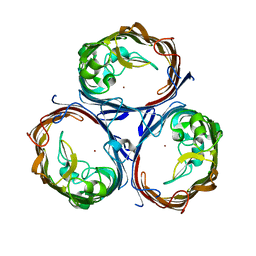 | | Structure of OmpF1 | | 分子名称: | Porin OmpF, ZINC ION | | 著者 | Jeong, W.J, Song, W.J. | | 登録日 | 2021-07-18 | | 公開日 | 2022-11-16 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (3.1 Å) | | 主引用文献 | Design and directed evolution of noncanonical beta-stereoselective metalloglycosidases.
Nat Commun, 13, 2022
|
|
7FF7
 
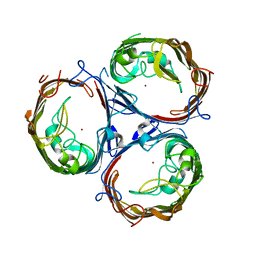 | | Structure of OmpF2 | | 分子名称: | Outer membrane protein F, ZINC ION | | 著者 | Jeong, W.J, Song, W.J. | | 登録日 | 2021-07-22 | | 公開日 | 2022-11-16 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (3.38 Å) | | 主引用文献 | Design and directed evolution of noncanonical beta-stereoselective metalloglycosidases.
Nat Commun, 13, 2022
|
|
4DTZ
 
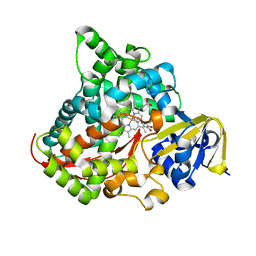 | | cytochrome P450 BM3h-8C8 MRI sensor bound to dopamine | | 分子名称: | L-DOPAMINE, PROTOPORPHYRIN IX CONTAINING FE, cytochrome P450 BM3 variant 8C8 | | 著者 | Brustad, E.M, Lelyveld, V.S, Snow, C.D, Crook, N, Martinez, F.M, Scholl, T.J, Jasanoff, A, Arnold, F.H. | | 登録日 | 2012-02-21 | | 公開日 | 2012-06-13 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | Structure-guided directed evolution of highly selective p450-based magnetic resonance imaging sensors for dopamine and serotonin.
J.Mol.Biol., 422, 2012
|
|
4DTY
 
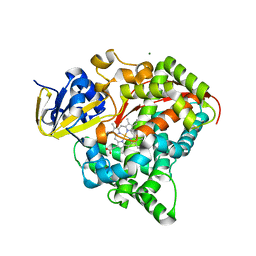 | | cytochrome P450 BM3h-8C8 MRI sensor, no ligand | | 分子名称: | MAGNESIUM ION, PROTOPORPHYRIN IX CONTAINING FE, cytochrome P450 BM3 variant 8C8 | | 著者 | Brustad, E.M, Lelyveld, V.S, Snow, C.D, Crook, N, Martinez, F.M, Scholl, T.J, Jasanoff, A, Arnold, F.H. | | 登録日 | 2012-02-21 | | 公開日 | 2012-06-13 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.45 Å) | | 主引用文献 | Structure-guided directed evolution of highly selective p450-based magnetic resonance imaging sensors for dopamine and serotonin.
J.Mol.Biol., 422, 2012
|
|
4DUC
 
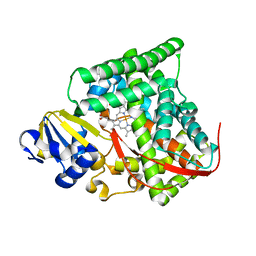 | | cytochrome P450 BM3h-2G9 MRI sensor, no ligand | | 分子名称: | PROTOPORPHYRIN IX CONTAINING FE, cytochrome P450 BM3 variant 2G9 | | 著者 | Brustad, E.M, Lelyveld, V.S, Snow, C.D, Crook, N, Martinez, F.M, Scholl, T.J, Jasanoff, A, Arnold, F.H. | | 登録日 | 2012-02-21 | | 公開日 | 2012-06-13 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.92 Å) | | 主引用文献 | Structure-guided directed evolution of highly selective p450-based magnetic resonance imaging sensors for dopamine and serotonin.
J.Mol.Biol., 422, 2012
|
|
4DUD
 
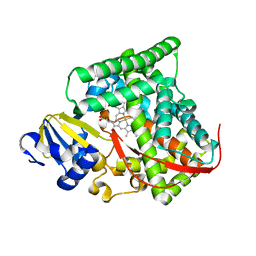 | | cytochrome P450 BM3h-2G9C6 MRI sensor, no ligand | | 分子名称: | PROTOPORPHYRIN IX CONTAINING FE, cytochrome P450 BM3 variant 2G9C6 | | 著者 | Brustad, E.M, Lelyveld, V.S, Snow, C.D, Crook, N, Martinez, F.M, Scholl, T.J, Jasanoff, A, Arnold, F.H. | | 登録日 | 2012-02-21 | | 公開日 | 2012-06-13 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Structure-guided directed evolution of highly selective p450-based magnetic resonance imaging sensors for dopamine and serotonin.
J.Mol.Biol., 422, 2012
|
|
4DUE
 
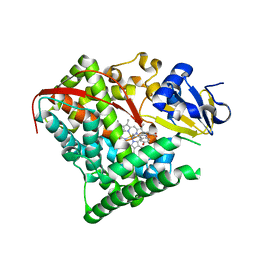 | | cytochrome P450 BM3h-2G9C6 MRI sensor bound to serotonin | | 分子名称: | PROTOPORPHYRIN IX CONTAINING FE, SEROTONIN, cytochrome P450 BM3 variant 2G9C6 | | 著者 | Brustad, E.M, Lelyveld, V.S, Snow, C.D, Crook, N, Martinez, F.M, Scholl, T.J, Jasanoff, A, Arnold, F.H. | | 登録日 | 2012-02-21 | | 公開日 | 2012-06-13 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Structure-guided directed evolution of highly selective p450-based magnetic resonance imaging sensors for dopamine and serotonin.
J.Mol.Biol., 422, 2012
|
|
4DTW
 
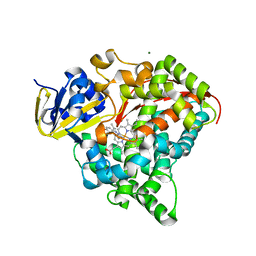 | | cytochrome P450 BM3h-8C8 MRI sensor bound to serotonin | | 分子名称: | Cytochrome P450 BM3 variant 8C8, MAGNESIUM ION, PROTOPORPHYRIN IX CONTAINING FE, ... | | 著者 | Brustad, E.M, Lelyveld, V.S, Snow, C.D, Crook, N, Martinez, F.M, Scholl, T.J, Jasanoff, A, Arnold, F.H. | | 登録日 | 2012-02-21 | | 公開日 | 2012-06-13 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structure-guided directed evolution of highly selective p450-based magnetic resonance imaging sensors for dopamine and serotonin.
J.Mol.Biol., 422, 2012
|
|
4DUA
 
 | | cytochrome P450 BM3h-9D7 MRI sensor, no ligand | | 分子名称: | PROTOPORPHYRIN IX CONTAINING FE, TRIETHYLENE GLYCOL, cytochrome P450 BM3 variant 9D7 | | 著者 | Brustad, E.M, Lelyveld, V.S, Snow, C.D, Crook, N, Martinez, F.M, Scholl, T.J, Jasanoff, A, Arnold, F.H. | | 登録日 | 2012-02-21 | | 公開日 | 2012-06-13 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structure-guided directed evolution of highly selective p450-based magnetic resonance imaging sensors for dopamine and serotonin.
J.Mol.Biol., 422, 2012
|
|
