1GRE
 
 | |
1GRH
 
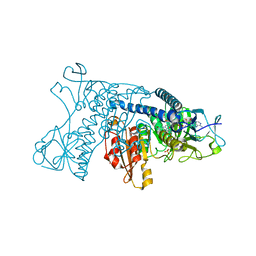 | | INHIBITION OF HUMAN GLUTATHIONE REDUCTASE BY THE NITROSOUREA DRUGS 1,3-BIS(2-CHLOROETHYL)-1-NITROSOUREA AND 1-(2-CHLOROETHYL)-3-(2-HYDROXYETHYL)-1-NITROSOUREA | | 分子名称: | ETHANOL, FLAVIN-ADENINE DINUCLEOTIDE, GLUTATHIONE REDUCTASE, ... | | 著者 | Karplus, P.A, Schulz, G.E. | | 登録日 | 1992-12-15 | | 公開日 | 1994-01-31 | | 最終更新日 | 2024-06-05 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Inhibition of human glutathione reductase by the nitrosourea drugs 1,3-bis(2-chloroethyl)-1-nitrosourea and 1-(2-chloroethyl)-3-(2-hydroxyethyl)-1-nitrosourea. A crystallographic analysis.
Eur.J.Biochem., 171, 1988
|
|
1GRA
 
 | |
1EAA
 
 | |
1EAE
 
 | |
1LVL
 
 | |
1EAC
 
 | |
1EAB
 
 | |
1EAD
 
 | |
1EAF
 
 | |
1AZB
 
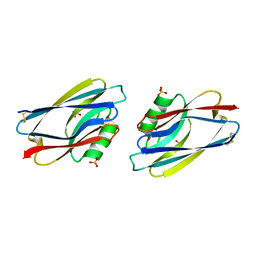 | |
1AZC
 
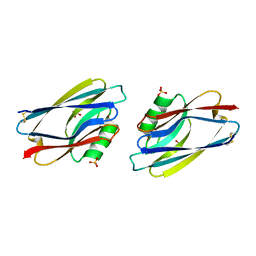 | | STRUCTURE OF APO-AZURIN FROM ALCALIGENES DENITRIFICANS AT 1.8 ANGSTROMS RESOLUTION | | 分子名称: | AZURIN, COPPER (II) ION, SULFATE ION | | 著者 | Baker, E.N, Shepard, W.E.B, Kingston, R.L. | | 登録日 | 1992-12-16 | | 公開日 | 1993-10-31 | | 最終更新日 | 2017-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structure of apo-azurin from Alcaligenes denitrificans at 1.8 A resolution.
Acta Crystallogr.,Sect.D, 49, 1993
|
|
1LED
 
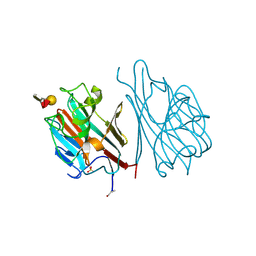 | | STRUCTURES OF THE LECTIN IV OF GRIFFONIA SIMPLICIFOLIA AND ITS COMPLEX WITH THE LEWIS B HUMAN BLOOD GROUP DETERMINANT AT 2.0 ANGSTROMS RESOLUTION | | 分子名称: | CALCIUM ION, MANGANESE (II) ION, SULFATE ION, ... | | 著者 | Delbaere, L, Vandonselaar, M, Quail, J. | | 登録日 | 1992-12-17 | | 公開日 | 1994-01-31 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structures of the lectin IV of Griffonia simplicifolia and its complex with the Lewis b human blood group determinant at 2.0 A resolution.
J.Mol.Biol., 230, 1993
|
|
1BRC
 
 | |
1LEC
 
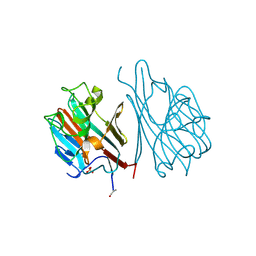 | | STRUCTURES OF THE LECTIN IV OF GRIFFONIA SIMPLICIFOLIA AND ITS COMPLEX WITH THE LEWIS B HUMAN BLOOD GROUP DETERMINANT AT 2.0 ANGSTROMS RESOLUTION | | 分子名称: | CALCIUM ION, MANGANESE (II) ION, SULFATE ION, ... | | 著者 | Delbaere, L, Vandonselaar, M, Quail, J. | | 登録日 | 1992-12-17 | | 公開日 | 1994-01-31 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structures of the lectin IV of Griffonia simplicifolia and its complex with the Lewis b human blood group determinant at 2.0 A resolution.
J.Mol.Biol., 230, 1993
|
|
1BRB
 
 | |
107L
 
 | |
108L
 
 | |
109L
 
 | |
112L
 
 | |
110L
 
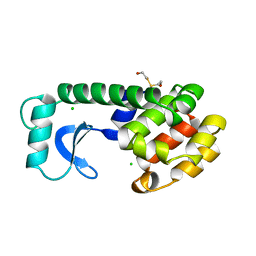 | |
111L
 
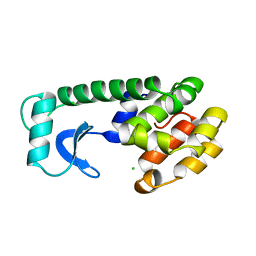 | |
113L
 
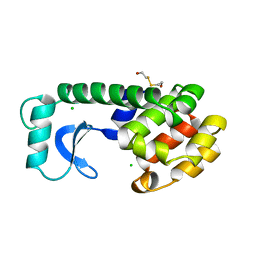 | |
115L
 
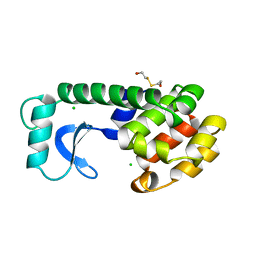 | |
114L
 
 | |
