7M50
 
 | |
1HVQ
 
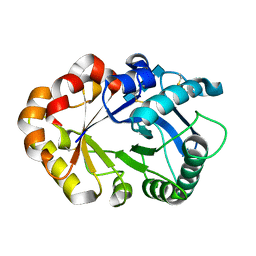 | | CRYSTAL STRUCTURES OF HEVAMINE, A PLANT DEFENCE PROTEIN WITH CHITINASE AND LYSOZYME ACTIVITY, AND ITS COMPLEX WITH AN INHIBITOR | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, HEVAMINE A | | 著者 | Terwisscha Van Scheltinga, A.C, Kalk, K.H, Beintema, J.J, Dijkstra, B.W. | | 登録日 | 1994-10-13 | | 公開日 | 1995-12-02 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Crystal structures of hevamine, a plant defence protein with chitinase and lysozyme activity, and its complex with an inhibitor.
Structure, 2, 1994
|
|
6APO
 
 | |
7MM8
 
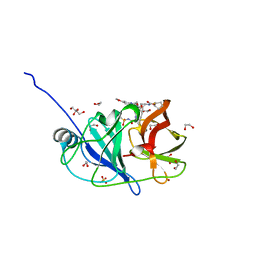 | | Crystal structure of HCV NS3/4A protease in complex with NR02-08 | | 分子名称: | (1R,2R)-2-fluorocyclopentyl {(2R,4S,6S,12Z,13aS,14aR,16aS)-2-[(7-methoxy-3-methylquinoxalin-2-yl)oxy]-14a-[(1-methylcyclopropane-1-sulfonyl)carbamoyl]-5,16-dioxo-1,2,3,5,6,7,8,9,10,11,13a,14,14a,15,16,16a-hexadecahydrocyclopropa[e]pyrrolo[1,2-a][1,4]diazacyclopentadecin-6-yl}carbamate, 1,2-ETHANEDIOL, NS3/4a protease, ... | | 著者 | Zephyr, J, Schiffer, C.A. | | 登録日 | 2021-04-29 | | 公開日 | 2022-03-09 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.43 Å) | | 主引用文献 | Deciphering the Molecular Mechanism of HCV Protease Inhibitor Fluorination as a General Approach to Avoid Drug Resistance.
J.Mol.Biol., 434, 2022
|
|
7MM6
 
 | |
7MMJ
 
 | | Crystal structure of HCV NS3/4A D168A protease in complex with NR02-08 | | 分子名称: | (1R,2R)-2-fluorocyclopentyl {(2R,4S,6S,12Z,13aS,14aR,16aS)-2-[(7-methoxy-3-methylquinoxalin-2-yl)oxy]-14a-[(1-methylcyclopropane-1-sulfonyl)carbamoyl]-5,16-dioxo-1,2,3,5,6,7,8,9,10,11,13a,14,14a,15,16,16a-hexadecahydrocyclopropa[e]pyrrolo[1,2-a][1,4]diazacyclopentadecin-6-yl}carbamate, 1,2-ETHANEDIOL, NS3 protease, ... | | 著者 | Zephyr, J, Schiffer, C.A. | | 登録日 | 2021-04-29 | | 公開日 | 2022-03-09 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.992 Å) | | 主引用文献 | Deciphering the Molecular Mechanism of HCV Protease Inhibitor Fluorination as a General Approach to Avoid Drug Resistance.
J.Mol.Biol., 434, 2022
|
|
7MM2
 
 | | Crystal structure of HCV NS3/4A protease in complex with NR02-61 | | 分子名称: | 1,2-ETHANEDIOL, 1-methylcyclobutyl [(2R,6S,12Z,13aS,14aR,16aS)-2-[(7-methoxy-3-methylquinoxalin-2-yl)oxy]-14a-{[(1-methylcyclopropyl)sulfonyl]carbamoyl}-5,16-dioxo-1,2,3,5,6,7,8,9,10,11,13a,14,14a,15,16,16a-hexadecahydrocyclopropa[e]pyrrolo[1,2-a][1,4]diazacyclopentadecin-6-yl]carbamate, NS3/4a protease, ... | | 著者 | Zephyr, J, Schiffer, C.A. | | 登録日 | 2021-04-29 | | 公開日 | 2022-03-09 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.891 Å) | | 主引用文献 | Deciphering the Molecular Mechanism of HCV Protease Inhibitor Fluorination as a General Approach to Avoid Drug Resistance.
J.Mol.Biol., 434, 2022
|
|
7MMF
 
 | | Crystal structure of HCV NS3/4A D168A protease in complex with NR02-60 | | 分子名称: | (1R)-1-cyclopentyl-2,2,2-trifluoroethyl {(2R,4R,6S,12Z,13aS,14aR,16aS)-2-[(7-methoxy-3-methylquinoxalin-2-yl)oxy]-14a-[(1-methylcyclopropane-1-sulfonyl)carbamoyl]-5,16-dioxo-1,2,3,5,6,7,8,9,10,11,13a,14,14a,15,16,16a-hexadecahydrocyclopropa[e]pyrrolo[1,2-a][1,4]diazacyclopentadecin-6-yl}carbamate, 1,2-ETHANEDIOL, NS3/4A protease, ... | | 著者 | Zephyr, J, Schiffer, C.A. | | 登録日 | 2021-04-29 | | 公開日 | 2022-03-09 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.891 Å) | | 主引用文献 | Deciphering the Molecular Mechanism of HCV Protease Inhibitor Fluorination as a General Approach to Avoid Drug Resistance.
J.Mol.Biol., 434, 2022
|
|
7MM7
 
 | | Crystal structure of HCV NS3/4A protease in complex with NR02-23 | | 分子名称: | (2S)-1,1,1-trifluoropropan-2-yl {(2R,4S,6S,12Z,13aS,14aR,16aS)-2-[(7-methoxy-3-methylquinoxalin-2-yl)oxy]-14a-[(1-methylcyclopropane-1-sulfonyl)carbamoyl]-5,16-dioxo-1,2,3,5,6,7,8,9,10,11,13a,14,14a,15,16,16a-hexadecahydrocyclopropa[e]pyrrolo[1,2-a][1,4]diazacyclopentadecin-6-yl}carbamate, 1,2-ETHANEDIOL, NS3/4a protease, ... | | 著者 | Zephyr, J, Schiffer, C.A. | | 登録日 | 2021-04-29 | | 公開日 | 2022-03-09 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.862 Å) | | 主引用文献 | Deciphering the Molecular Mechanism of HCV Protease Inhibitor Fluorination as a General Approach to Avoid Drug Resistance.
J.Mol.Biol., 434, 2022
|
|
7MMI
 
 | | Crystal structure of HCV NS3/4A D168A protease in complex with NR02-23 | | 分子名称: | (2S)-1,1,1-trifluoropropan-2-yl {(2R,4S,6S,12Z,13aS,14aR,16aS)-2-[(7-methoxy-3-methylquinoxalin-2-yl)oxy]-14a-[(1-methylcyclopropane-1-sulfonyl)carbamoyl]-5,16-dioxo-1,2,3,5,6,7,8,9,10,11,13a,14,14a,15,16,16a-hexadecahydrocyclopropa[e]pyrrolo[1,2-a][1,4]diazacyclopentadecin-6-yl}carbamate, NS3/4A protease, ZINC ION | | 著者 | Zephyr, J, Schiffer, C.A. | | 登録日 | 2021-04-29 | | 公開日 | 2022-03-09 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Deciphering the Molecular Mechanism of HCV Protease Inhibitor Fluorination as a General Approach to Avoid Drug Resistance.
J.Mol.Biol., 434, 2022
|
|
7MM5
 
 | | Crystal structure of HCV NS3/4A protease in complex with NR02-60 | | 分子名称: | (1R)-1-cyclopentyl-2,2,2-trifluoroethyl {(2R,4R,6S,12Z,13aS,14aR,16aS)-2-[(7-methoxy-3-methylquinoxalin-2-yl)oxy]-14a-[(1-methylcyclopropane-1-sulfonyl)carbamoyl]-5,16-dioxo-1,2,3,5,6,7,8,9,10,11,13a,14,14a,15,16,16a-hexadecahydrocyclopropa[e]pyrrolo[1,2-a][1,4]diazacyclopentadecin-6-yl}carbamate, 1,2-ETHANEDIOL, NS3/4a protease, ... | | 著者 | Zephyr, J, Schiffer, C.A. | | 登録日 | 2021-04-29 | | 公開日 | 2022-03-09 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.86 Å) | | 主引用文献 | Deciphering the Molecular Mechanism of HCV Protease Inhibitor Fluorination as a General Approach to Avoid Drug Resistance.
J.Mol.Biol., 434, 2022
|
|
7MML
 
 | | Crystal structure of HCV NS3/4A D168A protease in complex with NR01-145 | | 分子名称: | (2R)-1,1,1-trifluoropropan-2-yl {(2R,4R,6S,12Z,13aS,14aR,16aS)-2-[(7-methoxy-3-methylquinoxalin-2-yl)oxy]-14a-[(1-methylcyclopropane-1-sulfonyl)carbamoyl]-5,16-dioxo-1,2,3,5,6,7,8,9,10,11,13a,14,14a,15,16,16a-hexadecahydrocyclopropa[e]pyrrolo[1,2-a][1,4]diazacyclopentadecin-6-yl}carbamate, NS3 protease, SULFATE ION, ... | | 著者 | Zephyr, J, Schiffer, C.A. | | 登録日 | 2021-04-29 | | 公開日 | 2022-03-16 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.701 Å) | | 主引用文献 | Deciphering the Molecular Mechanism of HCV Protease Inhibitor Fluorination as a General Approach to Avoid Drug Resistance.
J.Mol.Biol., 434, 2022
|
|
7MMH
 
 | |
7MMK
 
 | | Crystal structure of HCV NS3/4A D168A protease in complex with NR01-149 | | 分子名称: | 1,2-ETHANEDIOL, 3,3-difluorocyclobutyl {(2R,4S,6S,12Z,13aS,14aR,16aS)-2-[(7-methoxy-3-methylquinoxalin-2-yl)oxy]-14a-[(1-methylcyclopropane-1-sulfonyl)carbamoyl]-5,16-dioxo-1,2,3,5,6,7,8,9,10,11,13a,14,14a,15,16,16a-hexadecahydrocyclopropa[e]pyrrolo[1,2-a][1,4]diazacyclopentadecin-6-yl}carbamate, NS3 protease, ... | | 著者 | Zephyr, J, Schiffer, C.A. | | 登録日 | 2021-04-29 | | 公開日 | 2022-03-16 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.89 Å) | | 主引用文献 | Deciphering the Molecular Mechanism of HCV Protease Inhibitor Fluorination as a General Approach to Avoid Drug Resistance.
J.Mol.Biol., 434, 2022
|
|
7MM9
 
 | | Crystal structure of HCV NS3/4A protease in complex with NR01-149 | | 分子名称: | 1,2-ETHANEDIOL, 3,3-difluorocyclobutyl {(2R,4S,6S,12Z,13aS,14aR,16aS)-2-[(7-methoxy-3-methylquinoxalin-2-yl)oxy]-14a-[(1-methylcyclopropane-1-sulfonyl)carbamoyl]-5,16-dioxo-1,2,3,5,6,7,8,9,10,11,13a,14,14a,15,16,16a-hexadecahydrocyclopropa[e]pyrrolo[1,2-a][1,4]diazacyclopentadecin-6-yl}carbamate, NS3 protease, ... | | 著者 | Zephyr, J, Schiffer, C.A. | | 登録日 | 2021-04-29 | | 公開日 | 2022-03-16 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.11 Å) | | 主引用文献 | Deciphering the Molecular Mechanism of HCV Protease Inhibitor Fluorination as a General Approach to Avoid Drug Resistance.
J.Mol.Biol., 434, 2022
|
|
7MMA
 
 | | Crystal structure of HCV NS3/4A protease in complex with NR01-145 | | 分子名称: | (2R)-1,1,1-trifluoropropan-2-yl {(2R,4R,6S,12Z,13aS,14aR,16aS)-2-[(7-methoxy-3-methylquinoxalin-2-yl)oxy]-14a-[(1-methylcyclopropane-1-sulfonyl)carbamoyl]-5,16-dioxo-1,2,3,5,6,7,8,9,10,11,13a,14,14a,15,16,16a-hexadecahydrocyclopropa[e]pyrrolo[1,2-a][1,4]diazacyclopentadecin-6-yl}carbamate, 1,2-ETHANEDIOL, NS3 protease, ... | | 著者 | Zephyr, J, Schiffer, C.A. | | 登録日 | 2021-04-29 | | 公開日 | 2022-03-16 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | Deciphering the Molecular Mechanism of HCV Protease Inhibitor Fluorination as a General Approach to Avoid Drug Resistance.
J.Mol.Biol., 434, 2022
|
|
7MM3
 
 | |
7MMB
 
 | |
7MMC
 
 | | Crystal structure of HCV NS3/4A D168A protease in complex with NR01-115 | | 分子名称: | (1-methylcyclopropyl)methyl {(2R,4S,6S,12Z,13aS,14aR,16aS)-2-[(7-methoxy-3-methylquinoxalin-2-yl)oxy]-14a-[(1-methylcyclopropane-1-sulfonyl)carbamoyl]-5,16-dioxo-1,2,3,5,6,7,8,9,10,11,13a,14,14a,15,16,16a-hexadecahydrocyclopropa[e]pyrrolo[1,2-a][1,4]diazacyclopentadecin-6-yl}carbamate, 1,2-ETHANEDIOL, NS3 protease, ... | | 著者 | Zephyr, J, Schiffer, C.A. | | 登録日 | 2021-04-29 | | 公開日 | 2022-03-16 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.001 Å) | | 主引用文献 | Deciphering the Molecular Mechanism of HCV Protease Inhibitor Fluorination as a General Approach to Avoid Drug Resistance.
J.Mol.Biol., 434, 2022
|
|
7MM4
 
 | | Crystal structure of HCV NS3/4A protease in complex with NR01-115 | | 分子名称: | (1-methylcyclopropyl)methyl {(2R,4S,6S,12Z,13aS,14aR,16aS)-2-[(7-methoxy-3-methylquinoxalin-2-yl)oxy]-14a-[(1-methylcyclopropane-1-sulfonyl)carbamoyl]-5,16-dioxo-1,2,3,5,6,7,8,9,10,11,13a,14,14a,15,16,16a-hexadecahydrocyclopropa[e]pyrrolo[1,2-a][1,4]diazacyclopentadecin-6-yl}carbamate, 1,2-ETHANEDIOL, NS3 protease, ... | | 著者 | Zephyr, J, Schiffer, C.A. | | 登録日 | 2021-04-29 | | 公開日 | 2022-03-16 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.89 Å) | | 主引用文献 | Deciphering the Molecular Mechanism of HCV Protease Inhibitor Fluorination as a General Approach to Avoid Drug Resistance.
J.Mol.Biol., 434, 2022
|
|
5FB6
 
 | | Room-temperature macromolecular crystallography using a micro-patterned silicon chip with minimal background scattering | | 分子名称: | Insulin Chain A, Insulin Chain B | | 著者 | Roedig, P, Duman, R, Sanchez-Weatherby, J, Vartiainen, I, Burkhardt, A, Warmer, M, David, C, Wagner, A, Meents, A. | | 登録日 | 2015-12-14 | | 公開日 | 2016-06-15 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.901 Å) | | 主引用文献 | Room-temperature macromolecular crystallography using a micro-patterned silicon chip with minimal background scattering.
J.Appl.Crystallogr., 49, 2016
|
|
7MLE
 
 | | Crystal Structure of HLA-A*03:01 in complex with VVRPSVASK, an 9-mer epitope from Influenza B virus | | 分子名称: | Beta-2-microglobulin, HLA class I histocompatibility antigen, A alpha chain, ... | | 著者 | Gras, S, Nguyen, A.T, Szeto, C. | | 登録日 | 2021-04-28 | | 公開日 | 2022-04-06 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Homologous peptides derived from influenza A, B and C viruses induce variable CD8 + T cell responses with cross-reactive potential.
Clin Transl Immunology, 11, 2022
|
|
2XPG
 
 | | Crystal structure of a MHC class I-peptide complex | | 分子名称: | BETA-2-MICROGLOBULIN, HLA CLASS I HISTOCOMPATIBILITY ANTIGEN, A-3 ALPHA CHAIN, ... | | 著者 | McMahon, R.M, Friis, L, Siebold, C, Friese, M.A, Fugger, L, Jones, E.Y. | | 登録日 | 2010-08-26 | | 公開日 | 2011-04-27 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Structure of Hla-A0301 in Complex with a Peptide of Proteolipid Protein: Insights Into the Role of Hla-A Alleles in Susceptibility to Multiple Sclerosis
Acta Crystallogr.,Sect.D, 67, 2011
|
|
7MMD
 
 | |
5E2Z
 
 | |
