8XC1
 
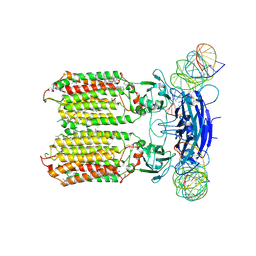 | | C. elegans SID1 in complex with dsRNA | | 分子名称: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Gong, D.S. | | 登録日 | 2023-12-07 | | 公開日 | 2024-06-05 | | 最終更新日 | 2024-07-03 | | 実験手法 | ELECTRON MICROSCOPY (2.21 Å) | | 主引用文献 | Structural basis for double-stranded RNA recognition by SID1.
Nucleic Acids Res., 52, 2024
|
|
8XBY
 
 | | The cryo-EM structure of the RAD51 L1 and L2 loops bound to the linker DNA with the blunt end of the nucleosome | | 分子名称: | DNA (5'-D(P*AP*AP*CP*GP*AP*AP*AP*AP*CP*GP*GP*CP*CP*AP*CP*CP*AP*CP*G)-3'), DNA (5'-D(P*CP*GP*TP*GP*GP*TP*GP*GP*CP*CP*GP*TP*TP*TP*TP*CP*GP*TP*T)-3'), DNA repair protein RAD51 homolog 1 | | 著者 | Shioi, T, Hatazawa, S, Ogasawara, M, Takizawa, Y, Kurumizaka, H. | | 登録日 | 2023-12-07 | | 公開日 | 2024-03-27 | | 最終更新日 | 2024-05-08 | | 実験手法 | ELECTRON MICROSCOPY (7.8 Å) | | 主引用文献 | Cryo-EM structures of RAD51 assembled on nucleosomes containing a DSB site.
Nature, 628, 2024
|
|
8XBX
 
 | | The cryo-EM structure of the RAD51 L2 loop bound to the linker DNA with the blunt end of the nucleosome | | 分子名称: | DNA (5'-D(P*AP*AP*CP*GP*AP*AP*AP*AP*CP*GP*GP*CP*CP*AP*CP*CP*AP*CP*G)-3'), DNA (5'-D(P*CP*GP*TP*GP*GP*TP*GP*GP*CP*CP*GP*TP*TP*TP*TP*CP*GP*TP*T)-3'), DNA repair protein RAD51 homolog 1 | | 著者 | Shioi, T, Hatazawa, S, Ogasawara, M, Takizawa, Y, Kurumizaka, H. | | 登録日 | 2023-12-07 | | 公開日 | 2024-03-27 | | 最終更新日 | 2024-04-17 | | 実験手法 | ELECTRON MICROSCOPY (4.36 Å) | | 主引用文献 | Cryo-EM structures of RAD51 assembled on nucleosomes containing a DSB site.
Nature, 628, 2024
|
|
8XBW
 
 | | The cryo-EM structure of the RAD51 N-terminal lobe domain bound to the histone H4 tail of the nucleosome | | 分子名称: | DNA (5'-D(P*AP*CP*CP*GP*CP*TP*TP*AP*AP*AP*CP*GP*CP*AP*CP*GP*TP*A)-3'), DNA (5'-D(P*TP*AP*CP*GP*TP*GP*CP*GP*TP*TP*TP*AP*AP*GP*CP*GP*GP*T)-3'), DNA repair protein RAD51 homolog 1, ... | | 著者 | Shioi, T, Hatazawa, S, Ogasawara, M, Takizawa, Y, Kurumizaka, H. | | 登録日 | 2023-12-07 | | 公開日 | 2024-03-27 | | 最終更新日 | 2024-05-08 | | 実験手法 | ELECTRON MICROSCOPY (2.89 Å) | | 主引用文献 | Cryo-EM structures of RAD51 assembled on nucleosomes containing a DSB site.
Nature, 628, 2024
|
|
8XBV
 
 | | The cryo-EM structure of the RAD51 L1 and L2 loops bound to the linker DNA with the sticky end of the nucleosome | | 分子名称: | DNA (5'-D(P*CP*GP*AP*AP*AP*AP*CP*GP*GP*CP*CP*AP*CP*CP*A)-3'), DNA (5'-D(P*TP*GP*GP*CP*CP*GP*TP*TP*TP*TP*CP*G)-3'), DNA repair protein RAD51 homolog 1 | | 著者 | Shioi, T, Hatazawa, S, Ogasawara, M, Takizawa, Y, Kurumizaka, H. | | 登録日 | 2023-12-07 | | 公開日 | 2024-03-27 | | 最終更新日 | 2024-05-08 | | 実験手法 | ELECTRON MICROSCOPY (7.61 Å) | | 主引用文献 | Cryo-EM structures of RAD51 assembled on nucleosomes containing a DSB site.
Nature, 628, 2024
|
|
8XBU
 
 | | The cryo-EM structure of the decameric RAD51 ring bound to the nucleosome with the linker DNA binding | | 分子名称: | DNA (153-MER), DNA (156-MER), DNA repair protein RAD51 homolog 1, ... | | 著者 | Shioi, T, Hatazawa, S, Ogasawara, M, Takizawa, Y, Kurumizaka, H. | | 登録日 | 2023-12-07 | | 公開日 | 2024-03-27 | | 最終更新日 | 2024-05-08 | | 実験手法 | ELECTRON MICROSCOPY (4.24 Å) | | 主引用文献 | Cryo-EM structures of RAD51 assembled on nucleosomes containing a DSB site.
Nature, 628, 2024
|
|
8XBT
 
 | | The cryo-EM structure of the octameric RAD51 ring bound to the nucleosome with the linker DNA binding | | 分子名称: | DNA (153-MER), DNA (156-MER), DNA repair protein RAD51 homolog 1, ... | | 著者 | Shioi, T, Hatazawa, S, Ogasawara, M, Takizawa, Y, Kurumizaka, H. | | 登録日 | 2023-12-07 | | 公開日 | 2024-03-27 | | 最終更新日 | 2024-05-08 | | 実験手法 | ELECTRON MICROSCOPY (4.12 Å) | | 主引用文献 | Cryo-EM structures of RAD51 assembled on nucleosomes containing a DSB site.
Nature, 628, 2024
|
|
8XBS
 
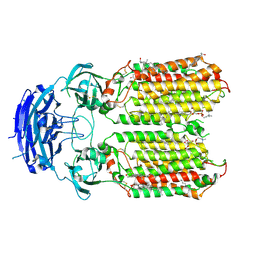 | | C. elegans apo-SID1 structure | | 分子名称: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Gong, D.S. | | 登録日 | 2023-12-07 | | 公開日 | 2024-06-05 | | 最終更新日 | 2024-07-03 | | 実験手法 | ELECTRON MICROSCOPY (2.21 Å) | | 主引用文献 | Structural basis for double-stranded RNA recognition by SID1.
Nucleic Acids Res., 52, 2024
|
|
8XBI
 
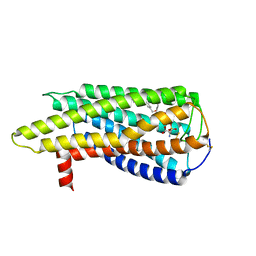 | | Human GPR34 -Gi complex bound to M1, receptor focused | | 分子名称: | (2~{S})-2-azanyl-3-[[(2~{R})-1-ethoxy-3-[3-[2-[(3-phenoxyphenyl)methoxy]phenyl]propanoyloxy]propan-2-yl]oxy-oxidanyl-phosphoryl]oxy-propanoic acid, Probable G-protein coupled receptor 34 | | 著者 | Kawahara, R, Shihoya, W, Nureki, O. | | 登録日 | 2023-12-06 | | 公開日 | 2023-12-27 | | 最終更新日 | 2024-05-15 | | 実験手法 | ELECTRON MICROSCOPY (3.06 Å) | | 主引用文献 | Structural basis for lysophosphatidylserine recognition by GPR34.
Nat Commun, 15, 2024
|
|
8XBH
 
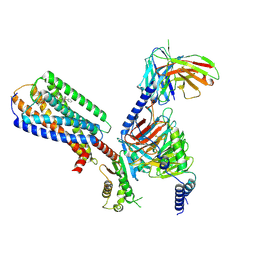 | | Human GPR34 -Gi complex bound to M1 | | 分子名称: | (2~{S})-2-azanyl-3-[[(2~{R})-1-ethoxy-3-[3-[2-[(3-phenoxyphenyl)methoxy]phenyl]propanoyloxy]propan-2-yl]oxy-oxidanyl-phosphoryl]oxy-propanoic acid, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | 著者 | Kawahara, R, Shihoya, W, Nureki, O. | | 登録日 | 2023-12-06 | | 公開日 | 2023-12-27 | | 最終更新日 | 2024-05-15 | | 実験手法 | ELECTRON MICROSCOPY (2.83 Å) | | 主引用文献 | Structural basis for lysophosphatidylserine recognition by GPR34.
Nat Commun, 15, 2024
|
|
8XBG
 
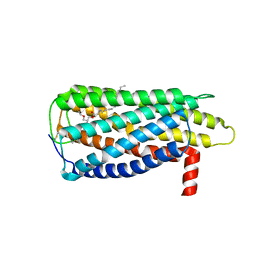 | |
8XBF
 
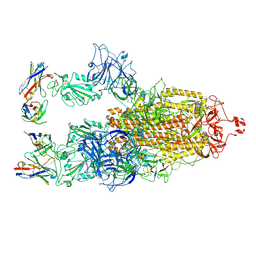 | | Cryo-EM structure of SARS-CoV-2 S-BQ.1 in complex with antibody O5C2 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, O5C2, heavy chain, ... | | 著者 | Hsu, H.F, Wu, M.H, Chang, Y.C, Hsu, S.T.D. | | 登録日 | 2023-12-06 | | 公開日 | 2024-06-19 | | 最終更新日 | 2024-06-26 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | Functional and structural investigation of a broadly neutralizing SARS-CoV-2 antibody.
JCI Insight, 9, 2024
|
|
8XBE
 
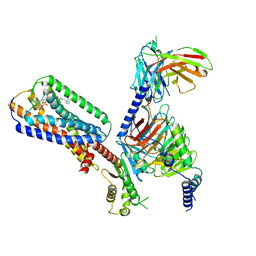 | | Human GPR34 -Gi complex bound to S3E-LysoPS | | 分子名称: | (2~{S})-2-azanyl-3-[[(2~{R})-1-ethoxy-3-[(~{Z})-octadec-9-enoyl]oxy-propan-2-yl]oxy-oxidanyl-phosphoryl]oxy-propanoic acid, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | 著者 | Kawahara, R, Shihoya, W, Nureki, O. | | 登録日 | 2023-12-06 | | 公開日 | 2024-05-15 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Structural basis for lysophosphatidylserine recognition by GPR34.
Nat Commun, 15, 2024
|
|
8XBD
 
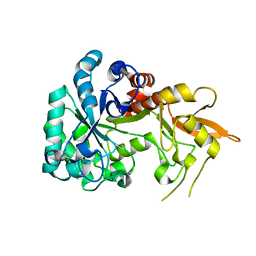 | |
8XB9
 
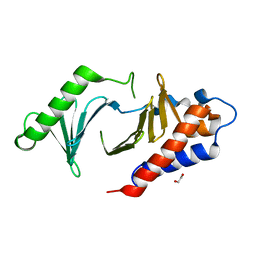 | | The Crystal Structure of polo-box domain of PLK1 from Biortus. | | 分子名称: | 1,2-ETHANEDIOL, Serine/threonine-protein kinase PLK1 | | 著者 | Wang, F, Cheng, W, Yuan, Z, Meng, Q, Zhang, B. | | 登録日 | 2023-12-06 | | 公開日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | The Crystal Structure of polo-box domain of PLK1 from Biortus
To Be Published
|
|
8XB8
 
 | |
8XB4
 
 | |
8XB3
 
 | |
8XB2
 
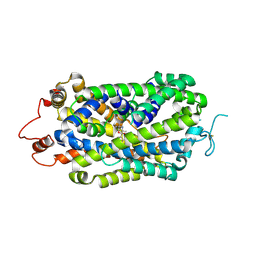 | | Structure of radafaxine-bound state of the human Norepinephrine Transporter | | 分子名称: | (2~{S},3~{S})-2-(3-chlorophenyl)-3,5,5-trimethyl-morpholin-2-ol, 2-acetamido-2-deoxy-beta-D-glucopyranose, GFP-MBP-solute carrier family 6 member 2,Maltose/maltodextrin-binding periplasmic protein,Sodium-dependent noradrenaline transporter,Maltose/maltodextrin-binding periplasmic protein,Sodium-dependent noradrenaline transporter | | 著者 | Wu, J.X, Ji, W.M. | | 登録日 | 2023-12-05 | | 公開日 | 2024-07-10 | | 実験手法 | ELECTRON MICROSCOPY (3.04 Å) | | 主引用文献 | Substrate binding and inhibition mechanism of norepinephrine transporter
To Be Published
|
|
8XB0
 
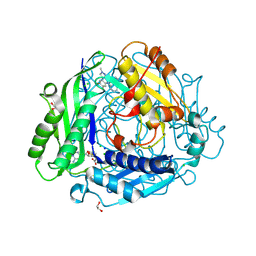 | | Structure-Based Design and Optimization of Methionine Adenosyltransferase 2A (MAT2A) Inhibitors with SAM and Compound 292 | | 分子名称: | 1,2-ETHANEDIOL, 7-chloranyl-5-(2-cyclopropylpyridin-3-yl)-8-fluoranyl-2-methyl-pyrazolo[3,4-c]quinolin-4-one, CHLORIDE ION, ... | | 著者 | Tong, S.L, Zhang, G.P. | | 登録日 | 2023-12-05 | | 公開日 | 2024-06-26 | | 実験手法 | X-RAY DIFFRACTION (1.12 Å) | | 主引用文献 | Structure-Based Design and Optimization of Methionine Adenosyltransferase 2A (MAT2A) Inhibitors with High Selectivity, Brain Penetration, and In Vivo Efficacy.
J.Med.Chem., 67, 2024
|
|
8XAY
 
 | |
8XAX
 
 | |
8XAW
 
 | |
8XAV
 
 | |
8XAU
 
 | | Cryo-EM structure of HerA | | 分子名称: | ATP-binding protein | | 著者 | Wang, Y, Deng, Z. | | 登録日 | 2023-12-05 | | 公開日 | 2024-06-05 | | 最終更新日 | 2024-07-10 | | 実験手法 | ELECTRON MICROSCOPY (3.14 Å) | | 主引用文献 | Molecular and structural basis of an ATPase-nuclease dual-enzyme anti-phage defense complex.
Cell Res., 2024
|
|
