5SPH
 
 | |
2RHK
 
 | | Crystal structure of influenza A NS1A protein in complex with F2F3 fragment of human cellular factor CPSF30, Northeast Structural Genomics Targets OR8C and HR6309A | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Cleavage and polyadenylation specificity factor subunit 4, NITRATE ION, ... | | 著者 | Das, K, Ma, L.-C, Xiao, R, Radvansky, B, Aramini, J, Zhao, L, Arnold, E, Krug, R.M, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | 登録日 | 2007-10-09 | | 公開日 | 2008-07-01 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Structural basis for suppression of a host antiviral response by influenza A virus.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
5SPK
 
 | |
2RMX
 
 | | Solution structure of the SHP-1 C-terminal SH2 domain complexed with a tyrosine-phosphorylated peptide from NKG2A | | 分子名称: | NKG2-A/NKG2-B type II integral membrane protein, Tyrosine-protein phosphatase non-receptor type 6 | | 著者 | Kasai, T, Koshiba, S, Inoue, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | 登録日 | 2007-11-30 | | 公開日 | 2008-12-02 | | 最終更新日 | 2022-03-16 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structural basis for the recognition of the two NKG2A immunoreceptor tyrosine-based inhibitory motifs (ITIMs) by the C-terminal SH2 domain of protein tyrosine phosphatase SHP-1
To be Published
|
|
4V6X
 
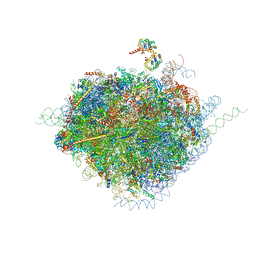 | | Structure of the human 80S ribosome | | 分子名称: | 18S ribosomal RNA, 28S ribosomal RNA, 40S ribosomal protein S10, ... | | 著者 | Anger, A.M, Armache, J.-P, Berninghausen, O, Habeck, M, Subklewe, M, Wilson, D.N, Beckmann, R. | | 登録日 | 2013-02-27 | | 公開日 | 2014-07-09 | | 最終更新日 | 2023-02-01 | | 実験手法 | ELECTRON MICROSCOPY (5 Å) | | 主引用文献 | Structures of the human and Drosophila 80S ribosome.
Nature, 497, 2013
|
|
8HS3
 
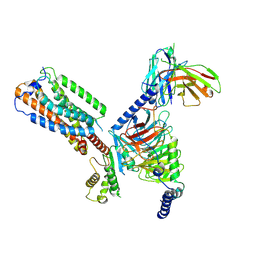 | | Gi bound orphan GPR20 in ligand-free state | | 分子名称: | Ggama, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | 著者 | Lin, X, Jiang, S, Xu, F. | | 登録日 | 2022-12-16 | | 公開日 | 2023-03-08 | | 最終更新日 | 2023-03-15 | | 実験手法 | ELECTRON MICROSCOPY (3.14 Å) | | 主引用文献 | The activation mechanism and antibody binding mode for orphan GPR20.
Cell Discov, 9, 2023
|
|
4USS
 
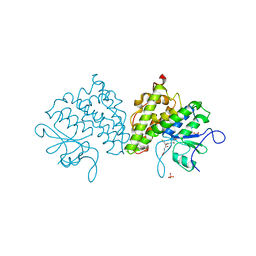 | | Populus trichocarpa glutathione transferase X1-1 (GHR1), complexed with glutathione | | 分子名称: | GLUTATHIONE, GLUTATHIONYL HYDROQUINONE REDUCTASE, PHOSPHATE ION | | 著者 | Lallement, P.A, Meux, E, Gualberto, J.M, Dumaracay, S, Favier, F, Didierjean, C, Saul, F, Haouz, A, Morel-Rouhier, M, Gelhaye, E, Rouhier, N, Hecker, A. | | 登録日 | 2014-07-13 | | 公開日 | 2014-12-03 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Glutathionyl-Hydroquinone Reductases from Poplar are Plastidial Proteins that Deglutathionylate Both Reduced and Oxidized Glutathionylated Quinones.
FEBS Lett., 589, 2015
|
|
8HFB
 
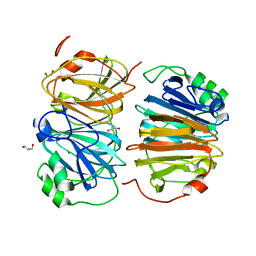 | | Evolved variant of quercetin 2,4-dioxygenase from Bacillus subtilis | | 分子名称: | 1,2-ETHANEDIOL, GLYCEROL, NICKEL (II) ION, ... | | 著者 | Eom, H, Song, W.J. | | 登録日 | 2022-11-10 | | 公開日 | 2023-03-08 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (2.24 Å) | | 主引用文献 | Underlying Role of Hydrophobic Environments in Tuning Metal Elements for Efficient Enzyme Catalysis.
J.Am.Chem.Soc., 145, 2023
|
|
5SPL
 
 | |
2RNO
 
 | |
4UT4
 
 | | Burkholderia pseudomallei heptokinase WcbL, D-mannose complex. | | 分子名称: | CHLORIDE ION, PUTATIVE SUGAR KINASE, alpha-D-mannopyranose | | 著者 | Vivoli, M, Isupov, M.N, Nicholas, R, Hill, A, Scott, A, Kosma, P, Prior, J, Harmer, N.J. | | 登録日 | 2014-07-18 | | 公開日 | 2016-01-13 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.94 Å) | | 主引用文献 | Unraveling the B.Pseudomallei Heptokinase Wcbl: From Structure to Drug Discovery.
Chem.Biol., 22, 2015
|
|
2RHX
 
 | |
5SPJ
 
 | |
5SPN
 
 | |
4UV2
 
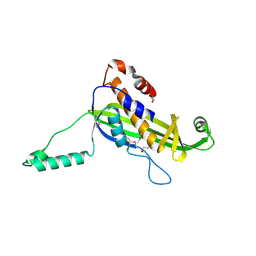 | | Structure of the curli transport lipoprotein CsgG in a non-lipidated, pre-pore conformation | | 分子名称: | CURLI PRODUCTION TRANSPORT COMPONENT CSGG | | 著者 | Goyal, P, Krasteva, P.V, Gerven, N.V, Gubellini, F, Broeck, I.V.D, Troupiotis-Tsailaki, A, Jonckheere, W, Pehau-Arnaudet, G, Pinkner, J.S, Chapman, M.R, Hultgren, S.J, Howorka, S, Fronzes, R, Remaut, H. | | 登録日 | 2014-08-04 | | 公開日 | 2014-09-24 | | 最終更新日 | 2014-12-17 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Structural and Mechanistic Insights Into the Bacterial Amyloid Secretion Channel Csgg.
Nature, 516, 2014
|
|
8HBA
 
 | |
2TPT
 
 | | STRUCTURAL AND THEORETICAL STUDIES SUGGEST DOMAIN MOVEMENT PRODUCES AN ACTIVE CONFORMATION OF THYMIDINE PHOSPHORYLASE | | 分子名称: | SULFATE ION, THYMIDINE PHOSPHORYLASE | | 著者 | Pugmire, M.J, Cook, W.J, Jasanoff, A, Walter, M.R, Ealick, S.E. | | 登録日 | 1997-11-24 | | 公開日 | 1999-03-02 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Structural and theoretical studies suggest domain movement produces an active conformation of thymidine phosphorylase.
J.Mol.Biol., 281, 1998
|
|
8HIL
 
 | | A cryo-EM structure of B. oleracea RNA polymerase V at 3.57 Angstrom | | 分子名称: | DNA-dependent RNA polymerase IV and V subunit 2, DNA-directed RNA polymerase V largest subunit, DNA-directed RNA polymerase subunit, ... | | 著者 | Du, X, Xie, G, Hu, H, Du, J. | | 登録日 | 2022-11-20 | | 公開日 | 2023-03-22 | | 最終更新日 | 2024-07-03 | | 実験手法 | ELECTRON MICROSCOPY (3.57 Å) | | 主引用文献 | Structure and mechanism of the plant RNA polymerase V.
Science, 379, 2023
|
|
5SPB
 
 | | PanDDA analysis group deposition -- Crystal structure of SARS-CoV-2 NSP3 macrodomain in complex with Z5010894404 - (R,R) and (S,S) isomers | | 分子名称: | (1R,2R)-4-hydroxy-1-[4-(phenylcarbamamido)benzamido]-2,3-dihydro-1H-indene-2-carboxylic acid, (1S,2S)-4-hydroxy-1-[4-(phenylcarbamamido)benzamido]-2,3-dihydro-1H-indene-2-carboxylic acid, Non-structural protein 3 | | 著者 | Correy, G.J, Fraser, J.S. | | 登録日 | 2022-06-09 | | 公開日 | 2022-07-13 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.05 Å) | | 主引用文献 | Iterative computational design and crystallographic screening identifies potent inhibitors targeting the Nsp3 macrodomain of SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
2RIG
 
 | |
4UXO
 
 | | Conserved mechanisms of microtubule-stimulated ADP release, ATP binding, and force generation in transport kinesins | | 分子名称: | GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, KINESIN-3 MOTOR DOMAIN, ... | | 著者 | Atherton, J, Farabella, I, Yu, I.M, Rosenfeld, S.S, Houdusse, A, Topf, M, Moores, C. | | 登録日 | 2014-08-27 | | 公開日 | 2014-09-24 | | 最終更新日 | 2024-05-08 | | 実験手法 | ELECTRON MICROSCOPY (6.3 Å) | | 主引用文献 | Conserved Mechanisms of Microtubule-Stimulated Adp Release, ATP Binding, and Force Generation in Transport Kinesins.
Elife, 3, 2014
|
|
4UZ5
 
 | |
8HBS
 
 | | Crystal of rAlfNmt | | 分子名称: | Glycylpeptide N-tetradecanoyltransferase | | 著者 | Wang, Y, Wang, S. | | 登録日 | 2022-10-30 | | 公開日 | 2023-03-08 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.78 Å) | | 主引用文献 | N -Myristoyltransferase, a Potential Antifungal Candidate Drug-Target for Aspergillus flavus.
Microbiol Spectr, 11, 2023
|
|
2ROB
 
 | | Solution structure of calcium bound soybean calmodulin isoform 4 C-terminal domain | | 分子名称: | CALCIUM ION, Calmodulin | | 著者 | Ishida, H, Huang, H, Yamniuk, A.P, Takaya, Y, Vogel, H.J. | | 登録日 | 2008-03-14 | | 公開日 | 2008-04-08 | | 最終更新日 | 2024-05-29 | | 実験手法 | SOLUTION NMR | | 主引用文献 | The solution structures of two soybean calmodulin isoforms provide a structural basis for their selective target activation properties
J.Biol.Chem., 283, 2008
|
|
5SOL
 
 | |
