3U5Z
 
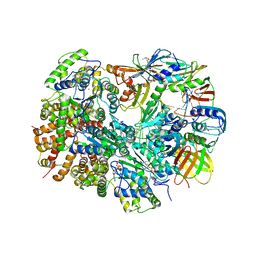 | | Structure of T4 Bacteriophage clamp loader bound to the T4 clamp, primer-template DNA, and ATP analog | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, DNA polymerase accessory protein 44, DNA polymerase accessory protein 62, ... | | 著者 | Kelch, B.A, Makino, D.L, O'Donnell, M, Kuriyan, J. | | 登録日 | 2011-10-11 | | 公開日 | 2012-01-04 | | 実験手法 | X-RAY DIFFRACTION (3.5 Å) | | 主引用文献 | How a DNA polymerase clamp loader opens a sliding clamp.
Science, 334, 2011
|
|
5FRQ
 
 | |
1IQP
 
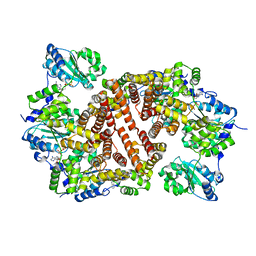 | | Crystal Structure of the Clamp Loader Small Subunit from Pyrococcus furiosus | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, RFCS | | 著者 | Oyama, T, Ishino, Y, Cann, I.K.O, Ishino, S, Morikawa, K. | | 登録日 | 2001-07-24 | | 公開日 | 2001-09-19 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Atomic Structure of the Clamp Loader Small Subunit from Pyrococcus furiosus
Mol.Cell, 8, 2001
|
|
3U60
 
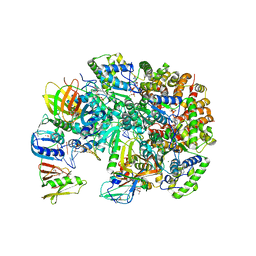 | | Structure of T4 Bacteriophage Clamp Loader Bound To Open Clamp, DNA and ATP Analog | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, DNA polymerase accessory protein 44, DNA polymerase accessory protein 62, ... | | 著者 | Kelch, B.A, Makino, D.L, O'Donnell, M, Kuriyan, J. | | 登録日 | 2011-10-11 | | 公開日 | 2012-01-04 | | 実験手法 | X-RAY DIFFRACTION (3.34 Å) | | 主引用文献 | How a DNA polymerase clamp loader opens a sliding clamp.
Science, 334, 2011
|
|
4TSZ
 
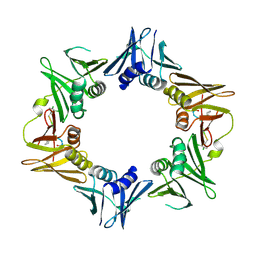 | | Crystal structure of DNA polymerase sliding clamp from Pseudomonas aeruginosa with ligand | | 分子名称: | ACE-GLN-ALC-ASP-LEU-ZCL peptide, DNA polymerase III subunit beta | | 著者 | Olieric, V, Burnouf, D, Ennifar, E, Wolff, P. | | 登録日 | 2014-06-19 | | 公開日 | 2014-09-10 | | 最終更新日 | 2016-12-21 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Differential Modes of Peptide Binding onto Replicative Sliding Clamps from Various Bacterial Origins.
J.Med.Chem., 57, 2014
|
|
8FS5
 
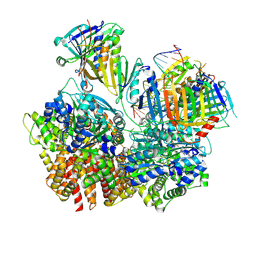 | | Structure of S. cerevisiae Rad24-RFC loading the 9-1-1 clamp onto a 10-nt gapped DNA in step 3 (open 9-1-1 and stably bound chamber DNA) | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, Checkpoint protein RAD24, DDC1 isoform 1, ... | | 著者 | Zheng, F, Georgescu, R, Yao, Y.N, O'Donnell, M.E, Li, H. | | 登録日 | 2023-01-09 | | 公開日 | 2023-06-14 | | 最終更新日 | 2024-06-19 | | 実験手法 | ELECTRON MICROSCOPY (2.76 Å) | | 主引用文献 | Structures of 9-1-1 DNA checkpoint clamp loading at gaps from start to finish and ramification to biology.
Biorxiv, 2023
|
|
8FS4
 
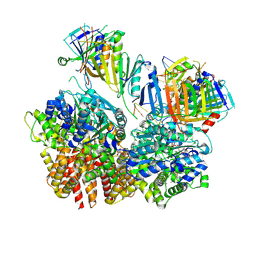 | | Structure of S. cerevisiae Rad24-RFC loading the 9-1-1 clamp onto a 10-nt gapped DNA in step 2 (open 9-1-1 ring and flexibly bound chamber DNA) | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, Checkpoint protein RAD24, DDC1 isoform 1, ... | | 著者 | Zheng, F, Georgescu, R, Yao, Y.N, O'Donnell, M.E, Li, H. | | 登録日 | 2023-01-09 | | 公開日 | 2023-06-14 | | 最終更新日 | 2024-06-19 | | 実験手法 | ELECTRON MICROSCOPY (2.94 Å) | | 主引用文献 | Structures of 9-1-1 DNA checkpoint clamp loading at gaps from start to finish and ramification to biology.
Biorxiv, 2023
|
|
8FS8
 
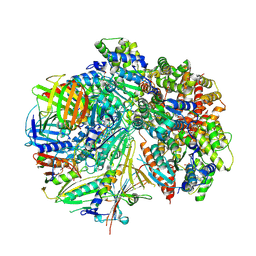 | | Structure of S. cerevisiae Rad24-RFC loading the 9-1-1 clamp onto a 5-nt gapped DNA (9-1-1 encircling fully bound DNA) | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, Checkpoint protein RAD24, DDC1 isoform 1, ... | | 著者 | Zheng, F, Georgescu, R, Yao, Y.N, O'Donnell, M.E, Li, H. | | 登録日 | 2023-01-09 | | 公開日 | 2023-06-14 | | 最終更新日 | 2024-06-19 | | 実験手法 | ELECTRON MICROSCOPY (3.04 Å) | | 主引用文献 | Structures of 9-1-1 DNA checkpoint clamp loading at gaps from start to finish and ramification to biology.
Biorxiv, 2023
|
|
8FS3
 
 | | Structure of S. cerevisiae Rad24-RFC loading the 9-1-1 clamp onto a 10-nt gapped DNA in step 1 (open 9-1-1 and shoulder bound DNA only) | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, Checkpoint protein RAD24, DDC1 isoform 1, ... | | 著者 | Zheng, F, Georgescu, R, Yao, Y.N, O'Donnell, M.E, Li, H. | | 登録日 | 2023-01-09 | | 公開日 | 2023-06-14 | | 最終更新日 | 2024-06-19 | | 実験手法 | ELECTRON MICROSCOPY (2.93 Å) | | 主引用文献 | Structures of 9-1-1 DNA checkpoint clamp loading at gaps from start to finish and ramification to biology.
Biorxiv, 2023
|
|
8FS6
 
 | | Structure of S. cerevisiae Rad24-RFC loading the 9-1-1 clamp onto a 10-nt gapped DNA in step 4 (partially closed 9-1-1 and stably bound chamber DNA) | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, Checkpoint protein RAD24, DDC1 isoform 1, ... | | 著者 | Zheng, F, Georgescu, R, Yao, Y.N, O'Donnell, M.E, Li, H. | | 登録日 | 2023-01-09 | | 公開日 | 2023-06-14 | | 最終更新日 | 2024-06-19 | | 実験手法 | ELECTRON MICROSCOPY (2.9 Å) | | 主引用文献 | Structures of 9-1-1 DNA checkpoint clamp loading at gaps from start to finish and ramification to biology.
Biorxiv, 2023
|
|
8FS7
 
 | | Structure of S. cerevisiae Rad24-RFC loading the 9-1-1 clamp onto a 10-nt gapped DNA in step 5 (closed 9-1-1 and stably bound chamber DNA) | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, Checkpoint protein RAD24, DDC1 isoform 1, ... | | 著者 | Zheng, F, Georgescu, R, Yao, Y.N, O'Donnell, M.E, Li, H. | | 登録日 | 2023-01-09 | | 公開日 | 2023-06-14 | | 最終更新日 | 2024-06-19 | | 実験手法 | ELECTRON MICROSCOPY (2.85 Å) | | 主引用文献 | Structures of 9-1-1 DNA checkpoint clamp loading at gaps from start to finish and ramification to biology.
Biorxiv, 2023
|
|
7NF9
 
 | | N-terminal C2H2 Zn-finger domain of Clamp | | 分子名称: | Chromatin-linked adaptor for MSL proteins, isoform A, ZINC ION | | 著者 | Mariasina, S.S, Bonchuk, A.N, Tikhonova, E.A, Efimov, S.V, Maksimenko, O.G, Georgiev, P.G, Polshakov, V.I. | | 登録日 | 2021-02-05 | | 公開日 | 2021-12-29 | | 最終更新日 | 2024-06-19 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structural basis for interaction between CLAMP and MSL2 proteins involved in the specific recruitment of the dosage compensation complex in Drosophila.
Nucleic Acids Res., 50, 2022
|
|
4YS0
 
 | |
6PTH
 
 | |
1UNN
 
 | |
5G4Q
 
 | | H.pylori Beta clamp in complex with 5-chloroisatin | | 分子名称: | 5-chloro-1H-indole-2,3-dione, DNA POLYMERASE III SUBUNIT BETA | | 著者 | Pandey, P, Gourinath, S. | | 登録日 | 2016-05-16 | | 公開日 | 2017-06-21 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Screening of E. coli beta-clamp Inhibitors Revealed that Few Inhibit Helicobacter pylori More Effectively: Structural and Functional Characterization.
Antibiotics (Basel), 7, 2018
|
|
3RB9
 
 | |
4TR7
 
 | |
4TR8
 
 | | Crystal structure of DNA polymerase sliding clamp from Pseudomonas aeruginosa | | 分子名称: | DNA polymerase III subunit beta, SODIUM ION | | 著者 | Olieric, V, Burnouf, D, Ennifar, E, Wolff, P. | | 登録日 | 2014-06-15 | | 公開日 | 2014-09-10 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Differential Modes of Peptide Binding onto Replicative Sliding Clamps from Various Bacterial Origins.
J.Med.Chem., 57, 2014
|
|
4TR6
 
 | | Crystal structure of DNA polymerase sliding clamp from Bacillus subtilis | | 分子名称: | DNA polymerase III subunit beta, SODIUM ION | | 著者 | Burnouf, D, Olieric, V, Ennifar, E, Wolff, P. | | 登録日 | 2014-06-14 | | 公開日 | 2014-09-10 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Differential Modes of Peptide Binding onto Replicative Sliding Clamps from Various Bacterial Origins.
J.Med.Chem., 57, 2014
|
|
1JR3
 
 | | Crystal Structure of the Processivity Clamp Loader Gamma Complex of E. coli DNA Polymerase III | | 分子名称: | DNA polymerase III subunit gamma, DNA polymerase III, delta subunit, ... | | 著者 | Jeruzalmi, D, O'Donnell, M, Kuriyan, J. | | 登録日 | 2001-08-10 | | 公開日 | 2001-09-26 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Crystal structure of the processivity clamp loader gamma (gamma) complex of E. coli DNA polymerase III.
Cell(Cambridge,Mass.), 106, 2001
|
|
5G48
 
 | | H.pylori Beta clamp in complex with Diflunisal | | 分子名称: | 5-(2,4-DIFLUOROPHENYL)-2-HYDROXY-BENZOIC ACID, DNA POLYMERASE III SUBUNIT BETA | | 著者 | Pandey, P, Gourinath, S. | | 登録日 | 2016-05-06 | | 公開日 | 2017-06-21 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.28 Å) | | 主引用文献 | Targeting the beta-clamp in Helicobacter pylori with FDA-approved drugs reveals micromolar inhibition by diflunisal.
FEBS Lett., 591, 2017
|
|
3F1V
 
 | | E. coli Beta Sliding Clamp, 148-153 Ala Mutant | | 分子名称: | CALCIUM ION, CHLORIDE ION, DNA polymerase III subunit beta | | 著者 | Cody, V. | | 登録日 | 2008-10-28 | | 公開日 | 2009-09-01 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.77 Å) | | 主引用文献 | Sliding clamp-DNA interactions are required for viability and contribute to DNA polymerase management in Escherichia coli.
J.Mol.Biol., 387, 2009
|
|
1NJF
 
 | | Nucleotide bound form of an isolated E. coli clamp loader gamma subunit | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, DNA polymerase III subunit gamma, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, ... | | 著者 | Podobnik, M, Weitze, T.F, O'Donnell, M, Kuriyan, J. | | 登録日 | 2002-12-30 | | 公開日 | 2003-04-08 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Nucleotide-Induced Conformational Changes in an Isolated Escherichia coli DNA Polymerase III Clamp Loader Subunit
Structure, 11, 2003
|
|
1NJG
 
 | | Nucleotide-free form of an Isolated E. coli Clamp Loader Gamma Subunit | | 分子名称: | DNA polymerase III subunit gamma, SULFATE ION, ZINC ION | | 著者 | Podobnik, M, Weitze, T.F, O'Donnell, M, Kuriyan, J. | | 登録日 | 2002-12-30 | | 公開日 | 2003-04-01 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Nucleotide-Induced Conformational Changes in an Isolated Escherichia coli DNA Polymerase III Clamp Loader Subunit
Structure, 11, 2003
|
|
