2JQ9
 
 | | VPS4A MIT-CHMP1A complex | | 分子名称: | Chromatin-modifying protein 1a, Vacuolar protein sorting-associating protein 4A | | 著者 | Stuchell-Brereton, M.D, Skalicky, J.J, Kieffer, C, Ghaffarian, S, Sundquist, W.I. | | 登録日 | 2007-05-30 | | 公開日 | 2007-10-16 | | 最終更新日 | 2023-12-20 | | 実験手法 | SOLUTION NMR | | 主引用文献 | ESCRT-III recognition by VPS4 ATPases.
Nature, 449, 2007
|
|
5MKY
 
 | | BROMODOMAIN OF HUMAN BRD9 WITH 4-chloro-2-methyl-5-((2-methyl-1,2,3,4-tetrahydroisoquinolin-5-yl)amino)pyridazin-3(2H)-one | | 分子名称: | 4-chloranyl-2-methyl-5-[(2-methyl-3,4-dihydro-1~{H}-isoquinolin-5-yl)amino]pyridazin-3-one, Bromodomain-containing protein 9 | | 著者 | Chung, C.-W. | | 登録日 | 2016-12-05 | | 公開日 | 2017-12-20 | | 最終更新日 | 2024-06-19 | | 実験手法 | X-RAY DIFFRACTION (1.67 Å) | | 主引用文献 | Discovery of a Potent, Cell Penetrant, and Selective p300/CBP-Associated Factor (PCAF)/General Control Nonderepressible 5 (GCN5) Bromodomain Chemical Probe.
J. Med. Chem., 60, 2017
|
|
4WQQ
 
 | | Structure of EPNH mutant of CEL-I | | 分子名称: | CALCIUM ION, Lectin CEL-I, N-acetyl-D-galactosamine-specific C-type, ... | | 著者 | Unno, H, Hatakeyama, T. | | 登録日 | 2014-10-22 | | 公開日 | 2015-04-29 | | 最終更新日 | 2024-11-06 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Mannose-recognition mutant of the galactose/N-acetylgalactosamine-specific C-type lectin CEL-I engineered by site-directed mutagenesis.
Biochim.Biophys.Acta, 1850, 2015
|
|
2C6C
 
 | | membrane-bound glutamate carboxypeptidase II (GCPII) in complex with GPI-18431 (S)-2-(4-iodobenzylphosphonomethyl)-pentanedioic acid | | 分子名称: | (2S)-2-{[HYDROXY(4-IODOBENZYL)PHOSPHORYL]METHYL}PENTANEDIOIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Mesters, J.R, Barinka, C, Li, W, Tsukamoto, T, Majer, P, Slusher, B.S, Konvalinka, J, Hilgenfeld, R. | | 登録日 | 2005-11-09 | | 公開日 | 2006-02-15 | | 最終更新日 | 2025-04-09 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structure of Glutamate Carboxypeptidase II, a Drug Target in Neuronal Damage and Prostate Cancer.
Embo J., 25, 2006
|
|
8D7K
 
 | | Bifunctional Inhibition of Neutrophil Elastase and Cathepsin G by Eap2 from S. aureus | | 分子名称: | Cathepsin G, C-terminal truncated form, Extracellular Adherence Protein, ... | | 著者 | Gido, C.D, Herdendorf, T.J, Geisbrecht, B.V. | | 登録日 | 2022-06-07 | | 公開日 | 2023-06-14 | | 最終更新日 | 2024-10-23 | | 実験手法 | X-RAY DIFFRACTION (3.1 Å) | | 主引用文献 | S. aureus Eap is a polyvalent inhibitor of neutrophil serine proteases.
J.Biol.Chem., 300, 2024
|
|
3V6N
 
 | |
7AZ6
 
 | | DNA polymerase sliding clamp from Escherichia coli with peptide 36 bound | | 分子名称: | ACETATE ION, Beta sliding clamp, CHLORIDE ION, ... | | 著者 | Monsarrat, C, Compain, G, Andre, C, Martiel, I, Engilberge, S, Olieric, V, Wolff, P, Brillet, K, Landolfo, M, Silva da Veiga, C, Wagner, J, Guichard, G, Burnouf, D.Y. | | 登録日 | 2020-11-16 | | 公開日 | 2021-12-01 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.93 Å) | | 主引用文献 | Iterative Structure-Based Optimization of Short Peptides Targeting the Bacterial Sliding Clamp.
J.Med.Chem., 64, 2021
|
|
1E7S
 
 | | GDP 4-keto-6-deoxy-D-mannose epimerase reductase K140R | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETYLPHOSPHATE, GDP-FUCOSE SYNTHETASE, ... | | 著者 | Rosano, C, Zuccotti, S, Izzo, G, Bolognesi, M. | | 登録日 | 2000-09-07 | | 公開日 | 2000-10-18 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Probing the Catalytic Mechanism of Gdp-4-Keto-6-Deoxy-D-Mannose Epimerase/Reductase by Kinetic and Crystallographic Characterization of Site-Specific Mutants
J.Mol.Biol., 303, 2000
|
|
7AZ8
 
 | | DNA polymerase sliding clamp from Escherichia coli with peptide 43 bound | | 分子名称: | Beta sliding clamp, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | 著者 | Monsarrat, C, Compain, G, Andre, C, Martiel, I, Engilberge, S, Olieric, V, Wolff, P, Brillet, K, Landolfo, M, Silva da Veiga, C, Wagner, J, Guichard, G, Burnouf, D.Y. | | 登録日 | 2020-11-16 | | 公開日 | 2021-12-01 | | 最終更新日 | 2024-11-06 | | 実験手法 | X-RAY DIFFRACTION (1.61 Å) | | 主引用文献 | Iterative Structure-Based Optimization of Short Peptides Targeting the Bacterial Sliding Clamp.
J.Med.Chem., 64, 2021
|
|
7AZE
 
 | | DNA polymerase sliding clamp from Escherichia coli with peptide 18 bound | | 分子名称: | Beta sliding clamp, GLYCEROL, MALONATE ION, ... | | 著者 | Monsarrat, C, Compain, G, Andre, C, Martiel, I, Engilberge, S, Olieric, V, Wolff, P, Brillet, K, Landolfo, M, Silva da Veiga, C, Wagner, J, Guichard, G, Burnouf, D.Y. | | 登録日 | 2020-11-16 | | 公開日 | 2021-12-01 | | 最終更新日 | 2024-10-23 | | 実験手法 | X-RAY DIFFRACTION (1.82 Å) | | 主引用文献 | Iterative Structure-Based Optimization of Short Peptides Targeting the Bacterial Sliding Clamp.
J.Med.Chem., 64, 2021
|
|
2JQU
 
 | |
8CK4
 
 | | STRUCTURE OF HIF2A-ARNT HETERODIMER IN COMPLEX WITH (4S)-1-(3,5-difluorophenyl)-5,5-difluoro-3-methanesulfonyl-4,5,6,7-tetrahydro-2-benzothiophen-4-ol | | 分子名称: | (4~{S})-1-[3,5-bis(fluoranyl)phenyl]-5,5-bis(fluoranyl)-3-methylsulfonyl-6,7-dihydro-4~{H}-2-benzothiophen-4-ol, Aryl hydrocarbon receptor nuclear translocator, Endothelial PAS domain-containing protein 1 | | 著者 | Musil, D. | | 登録日 | 2023-02-14 | | 公開日 | 2023-07-19 | | 最終更新日 | 2024-06-19 | | 実験手法 | X-RAY DIFFRACTION (2.29 Å) | | 主引用文献 | Discovery of Cycloalkyl[ c ]thiophenes as Novel Scaffolds for Hypoxia-Inducible Factor-2 alpha Inhibitors.
J.Med.Chem., 66, 2023
|
|
7XBZ
 
 | | Crystal structure of Staphylococcus aureus ClpP in complex with R-ZG197 | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, (6S,9aS)-6-[(2S)-butan-2-yl]-8-[(1R)-1-naphthalen-1-ylethyl]-4,7-bis(oxidanylidene)-N-[4,4,4-tris(fluoranyl)butyl]-3,6,9,9a-tetrahydro-2H-pyrazino[1,2-a]pyrimidine-1-carboxamide, ATP-dependent Clp protease proteolytic subunit, ... | | 著者 | Wei, B.Y, Gan, J.H, Yang, C.-G. | | 登録日 | 2022-03-22 | | 公開日 | 2022-11-16 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.15 Å) | | 主引用文献 | Anti-infective therapy using species-specific activators of Staphylococcus aureus ClpP.
Nat Commun, 13, 2022
|
|
8Z1F
 
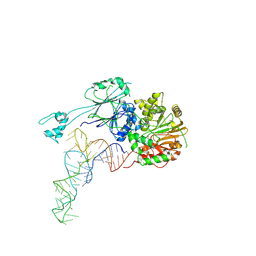 | | Cryo-EM structure of human ELAC2-tRNA | | 分子名称: | Homo sapiens mitochondrion pre-tRNA-Tyr, PHOSPHATE ION, ZINC ION, ... | | 著者 | Liu, Z.M, Xue, C.Y. | | 登録日 | 2024-04-11 | | 公開日 | 2024-12-18 | | 最終更新日 | 2025-06-25 | | 実験手法 | ELECTRON MICROSCOPY (4.3 Å) | | 主引用文献 | Structural insights into human ELAC2 as a tRNA 3' processing enzyme.
Nucleic Acids Res., 52, 2024
|
|
2JSZ
 
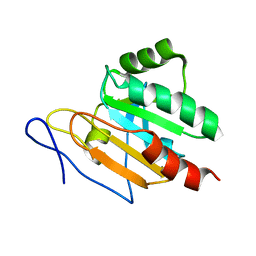 | |
4ZUR
 
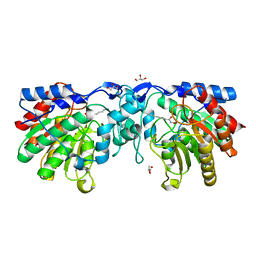 | | Crystal structure of acetylpolyamine amidohydrolase from Mycoplana ramosa in complex with a hydroxamate inhibitor | | 分子名称: | 7-amino-N-hydroxyheptanamide, AMMONIUM ION, Acetylpolyamine aminohydrolase, ... | | 著者 | Decroos, C, Christianson, D.W. | | 登録日 | 2015-05-17 | | 公開日 | 2015-07-29 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.13 Å) | | 主引用文献 | Design, Synthesis, and Evaluation of Polyamine Deacetylase Inhibitors, and High-Resolution Crystal Structures of Their Complexes with Acetylpolyamine Amidohydrolase.
Biochemistry, 54, 2015
|
|
6OGY
 
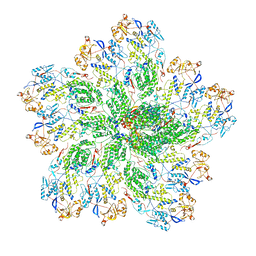 | | In situ structure of Rotavirus RNA-dependent RNA polymerase at duplex-open state | | 分子名称: | DNA/RNA (5'-D(*(GTG))-R(P*GP*C)-3'), Inner capsid protein VP2, RNA (5'-R(P*AP*GP*CP*C)-3'), ... | | 著者 | Ding, K, Chang, T, Shen, W, Roy, P, Zhou, Z.H. | | 登録日 | 2019-04-03 | | 公開日 | 2019-05-22 | | 最終更新日 | 2025-05-21 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | In situ structures of rotavirus polymerase in action and mechanism of mRNA transcription and release.
Nat Commun, 10, 2019
|
|
1HZN
 
 | |
2ANB
 
 | | Crystal Structure Of Oligomeric E.coli Guanylate Kinase In Complex With GMP | | 分子名称: | GUANOSINE-5'-MONOPHOSPHATE, Guanylate kinase, SULFATE ION | | 著者 | Hible, G, Renault, L, Schaeffer, F, Christova, P, Radulescu, A.Z, Evrin, C, Gilles, A.M, Cherfils, J. | | 登録日 | 2005-08-11 | | 公開日 | 2005-08-30 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Calorimetric and crystallographic analysis of the oligomeric structure of Escherichia coli GMP kinase
J.Mol.Biol., 352, 2005
|
|
3UXU
 
 | |
4JRU
 
 | | Structure of haze forming proteins in white wines: Vitis vinifera thaumatin-like proteins | | 分子名称: | GLYCEROL, thaumatin-like protein | | 著者 | Marangon, M, Menz, R.I, Waters, E.J, Van Sluyter, S.C. | | 登録日 | 2013-03-22 | | 公開日 | 2014-04-02 | | 最終更新日 | 2024-11-20 | | 実験手法 | X-RAY DIFFRACTION (1.2 Å) | | 主引用文献 | Structure of Haze Forming Proteins in White Wines: Vitis vinifera Thaumatin-Like Proteins.
Plos One, 9, 2014
|
|
2K2R
 
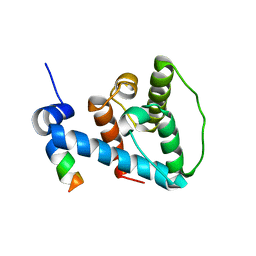 | | The NMR structure of alpha-parvin CH2/paxillin LD1 complex | | 分子名称: | Alpha-parvin, Paxillin | | 著者 | Wang, X, Fukuda, K, Byeon, I, Velyvis, A, Wu, C, Gronenborn, A, Qin, J. | | 登録日 | 2008-04-10 | | 公開日 | 2008-05-27 | | 最終更新日 | 2024-05-29 | | 実験手法 | SOLUTION NMR | | 主引用文献 | The Structure of {alpha}-Parvin CH2-Paxillin LD1 Complex Reveals a Novel Modular Recognition for Focal Adhesion Assembly.
J.Biol.Chem., 283, 2008
|
|
5A48
 
 | |
1FCA
 
 | |
5MKZ
 
 | |
