1B1B
 
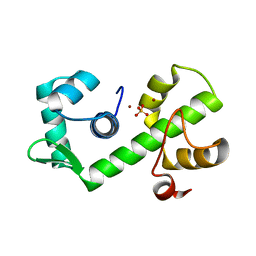 | | IRON DEPENDENT REGULATOR | | 分子名称: | PROTEIN (IRON DEPENDENT REGULATOR), SULFATE ION, ZINC ION | | 著者 | Pohl, E, Holmes, R.K, Hol, W.G. | | 登録日 | 1998-11-19 | | 公開日 | 1999-12-03 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Crystal structure of the iron-dependent regulator (IdeR) from Mycobacterium tuberculosis shows both metal binding sites fully occupied.
J.Mol.Biol., 285, 1999
|
|
1B09
 
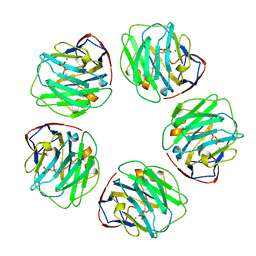 | |
1B22
 
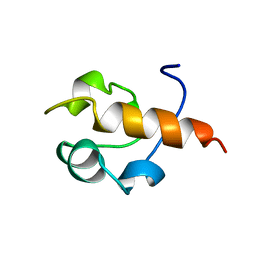 | | RAD51 (N-TERMINAL DOMAIN) | | 分子名称: | DNA REPAIR PROTEIN RAD51 | | 著者 | Aihara, H, Ito, Y, Kurumizaka, H, Yokoyama, S, Shibata, T, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | 登録日 | 1998-12-04 | | 公開日 | 1999-12-03 | | 最終更新日 | 2023-12-27 | | 実験手法 | SOLUTION NMR | | 主引用文献 | The N-terminal domain of the human Rad51 protein binds DNA: structure and a DNA binding surface as revealed by NMR.
J.Mol.Biol., 290, 1999
|
|
1B26
 
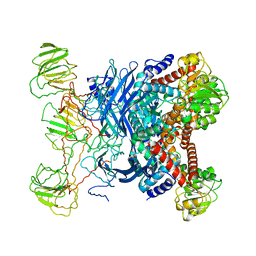 | | GLUTAMATE DEHYDROGENASE | | 分子名称: | GLUTAMATE DEHYDROGENASE | | 著者 | Knapp, S, Devos, W.M, Rice, D, Ladenstein, R. | | 登録日 | 1998-12-04 | | 公開日 | 1999-12-03 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Crystal structure of glutamate dehydrogenase from the hyperthermophilic eubacterium Thermotoga maritima at 3.0 A resolution.
J.Mol.Biol., 267, 1997
|
|
2TMG
 
 | | THERMOTOGA MARITIMA GLUTAMATE DEHYDROGENASE MUTANT S128R, T158E, N117R, S160E | | 分子名称: | PROTEIN (GLUTAMATE DEHYDROGENASE) | | 著者 | Knapp, S, Lebbink, J.H.G, van der Oost, J, de Vos, W.M, Rice, D, Ladenstein, R. | | 登録日 | 1998-12-04 | | 公開日 | 1999-12-08 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Engineering activity and stability of Thermotoga maritima glutamate dehydrogenase. II: construction of a 16-residue ion-pair network at the subunit interface.
J.Mol.Biol., 289, 1999
|
|
3RAP
 
 | |
1DI5
 
 | |
1DG4
 
 | | NMR STRUCTURE OF THE SUBSTRATE BINDING DOMAIN OF DNAK IN THE APO FORM | | 分子名称: | DNAK | | 著者 | Pellecchia, M, Montgomery, D.L, Stevens, S.Y, Van der Kooi, C.W, Feng, H, Gierasch, L.M, Zuiderweg, E.R.P. | | 登録日 | 1999-11-23 | | 公開日 | 1999-12-08 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structural insights into substrate binding by the molecular chaperone DnaK.
Nat.Struct.Biol., 7, 2000
|
|
1DII
 
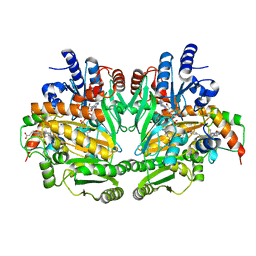 | | CRYSTAL STRUCTURE OF P-CRESOL METHYLHYDROXYLASE AT 2.5 A RESOLUTION | | 分子名称: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, HEME C, ... | | 著者 | Cunane, L.M, Chen, Z.W, Shamala, N, Mathews, F.S, Cronin, C.N, McIntire, W.S. | | 登録日 | 1999-11-29 | | 公開日 | 1999-12-08 | | 最終更新日 | 2021-03-03 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Structures of the flavocytochrome p-cresol methylhydroxylase and its enzyme-substrate complex: gated substrate entry and proton relays support the proposed catalytic mechanism.
J.Mol.Biol., 295, 2000
|
|
1DGU
 
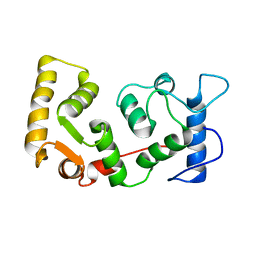 | |
1DGZ
 
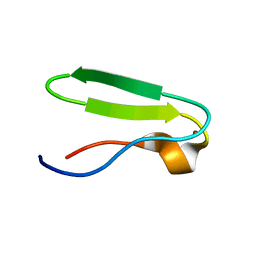 | | RIBOSMAL PROTEIN L36 FROM THERMUS THERMOPHILUS: NMR STRUCTURE ENSEMBLE | | 分子名称: | PROTEIN (L36 RIBOSOMAL PROTEIN), ZINC ION | | 著者 | Hard, T, Rak, A, Allard, P, Kloo, L, Garber, M. | | 登録日 | 1999-11-27 | | 公開日 | 1999-12-08 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | The solution structure of ribosomal protein L36 from Thermus thermophilus reveals a zinc-ribbon-like fold.
J.Mol.Biol., 296, 2000
|
|
1DIQ
 
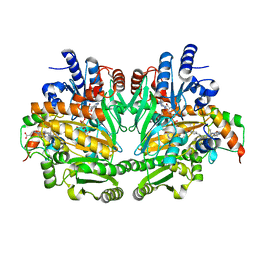 | | CRYSTAL STRUCTURE OF P-CRESOL METHYLHYDROXYLASE WITH SUBSTRATE BOUND | | 分子名称: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, HEME C, ... | | 著者 | Cunane, L.M, Chen, Z.W, Shamala, N, Mathews, F.S, Cronin, C.S, McIntire, W.S. | | 登録日 | 1999-11-29 | | 公開日 | 1999-12-08 | | 最終更新日 | 2021-03-03 | | 実験手法 | X-RAY DIFFRACTION (2.75 Å) | | 主引用文献 | Structures of the flavocytochrome p-cresol methylhydroxylase and its enzyme-substrate complex: gated substrate entry and proton relays support the proposed catalytic mechanism.
J.Mol.Biol., 295, 2000
|
|
1DGV
 
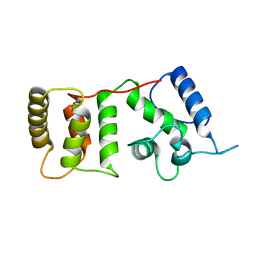 | |
1B3B
 
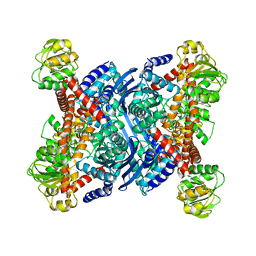 | | THERMOTOGA MARITIMA GLUTAMATE DEHYDROGENASE MUTANT N97D, G376K | | 分子名称: | PROTEIN (GLUTAMATE DEHYDROGENASE) | | 著者 | Knapp, S, Lebbink, J.H.G, Van Der Oost, J, Devos, W.M, Rice, D, Ladenstein, R. | | 登録日 | 1998-12-07 | | 公開日 | 1999-12-08 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (3.1 Å) | | 主引用文献 | Engineering activity and stability of Thermotoga maritima glutamate dehydrogenase. I. Introduction of a six-residue ion-pair network in the hinge region.
J.Mol.Biol., 280, 1998
|
|
1DI3
 
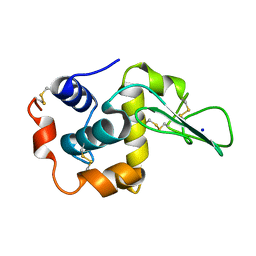 | |
1DF1
 
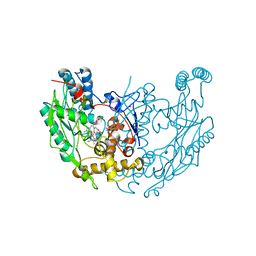 | | MURINE INOSOXY DIMER WITH ISOTHIOUREA BOUND IN THE ACTIVE SITE | | 分子名称: | 5,6,7,8-TETRAHYDROBIOPTERIN, ETHYLISOTHIOUREA, NITRIC OXIDE SYNTHASE, ... | | 著者 | Crane, B.R, Rosenfeld, R.J, Arvai, A.S, Ghosh, D.K, Ghosh, S, Tainer, J.A, Stuehr, D.J, Getzoff, E.D. | | 登録日 | 1999-11-16 | | 公開日 | 1999-12-08 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.35 Å) | | 主引用文献 | N-terminal domain swapping and metal ion binding in nitric oxide synthase dimerization.
EMBO J., 18, 1999
|
|
1DI4
 
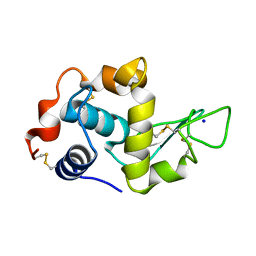 | |
1DG9
 
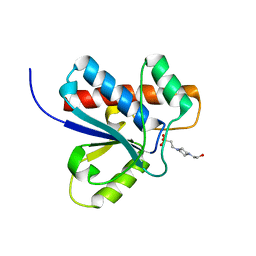 | | CRYSTAL STRUCTURE OF BOVINE LOW MOLECULAR WEIGHT PTPASE COMPLEXED WITH HEPES | | 分子名称: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, TYROSINE PHOSPHATASE | | 著者 | Zhang, M, Zhou, M, Van Etten, R.L, Stauffacher, C.V. | | 登録日 | 1999-11-23 | | 公開日 | 1999-12-08 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Crystal structure of bovine low molecular weight phosphotyrosyl phosphatase complexed with the transition state analog vanadate.
Biochemistry, 36, 1997
|
|
1CM5
 
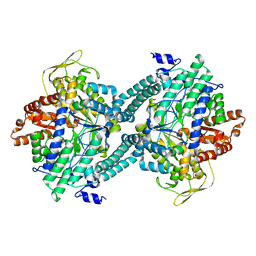 | | CRYSTAL STRUCTURE OF C418A,C419A MUTANT OF PFL FROM E.COLI | | 分子名称: | CARBONATE ION, PROTEIN (PYRUVATE FORMATE-LYASE), SODIUM ION | | 著者 | Becker, A, Fritz-Wolf, K, Kabsch, W, Knappe, J, Schultz, S, Wagner, A.F.V. | | 登録日 | 1999-05-14 | | 公開日 | 1999-12-08 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Structure and mechanism of the glycyl radical enzyme pyruvate formate-lyase.
Nat.Struct.Biol., 6, 1999
|
|
1B3D
 
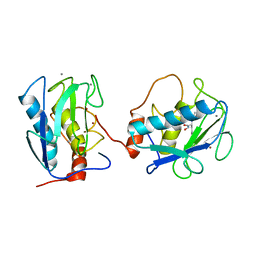 | | STROMELYSIN-1 | | 分子名称: | CALCIUM ION, N-[[2-METHYL-4-HYDROXYCARBAMOYL]BUT-4-YL-N]-BENZYL-P-[PHENYL]-P-[METHYL]PHOSPHINAMID, STROMELYSIN-1, ... | | 著者 | Chen, L, Rydel, T.J, Dunaway, C.M, Pikul, S, Dunham, K.M, Gu, F, Barnett, B.L. | | 登録日 | 1998-12-09 | | 公開日 | 1999-12-10 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Crystal structure of the stromelysin catalytic domain at 2.0 A resolution: inhibitor-induced conformational changes.
J.Mol.Biol., 293, 1999
|
|
1D1D
 
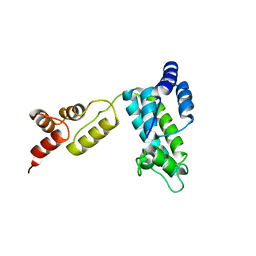 | |
1DJF
 
 | | NMR STRUCTURE OF A MODEL HYDROPHILIC AMPHIPATHIC HELICAL BASIC PEPTIDE | | 分子名称: | GLN-ALA-PRO-ALA-TYR-LYS-LYS-ALA-ALA-LYS-LYS-LEU-ALA-GLU-SER | | 著者 | Montserret, R, McLeish, M.J, Bockmann, A, Geourjon, C, Penin, F. | | 登録日 | 1999-12-03 | | 公開日 | 1999-12-10 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Involvement of electrostatic interactions in the mechanism of peptide folding induced by sodium dodecyl sulfate binding.
Biochemistry, 39, 2000
|
|
1DJ8
 
 | |
1DFO
 
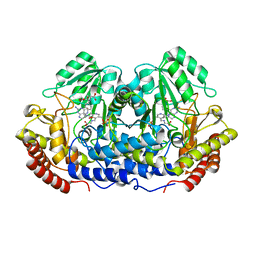 | | CRYSTAL STRUCTURE AT 2.4 ANGSTROM RESOLUTION OF E. COLI SERINE HYDROXYMETHYLTRANSFERASE IN COMPLEX WITH GLYCINE AND 5-FORMYL TETRAHYDROFOLATE | | 分子名称: | N-GLYCINE-[3-HYDROXY-2-METHYL-5-PHOSPHONOOXYMETHYL-PYRIDIN-4-YL-METHANE], N-[4-({[(6S)-2-amino-5-formyl-4-oxo-3,4,5,6,7,8-hexahydropteridin-6-yl]methyl}amino)benzoyl]-L-glutamic acid, SERINE HYDROXYMETHYLTRANSFERASE | | 著者 | Scarsdale, J.N, Radaev, S, Kazanina, G, Schirch, V, Wright, H.T. | | 登録日 | 1999-11-20 | | 公開日 | 1999-12-10 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Crystal structure at 2.4 A resolution of E. coli serine hydroxymethyltransferase in complex with glycine substrate and 5-formyl tetrahydrofolate.
J.Mol.Biol., 296, 2000
|
|
1QR0
 
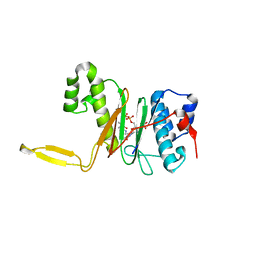 | | CRYSTAL STRUCTURE OF THE 4'-PHOSPHOPANTETHEINYL TRANSFERASE SFP-COENZYME A COMPLEX | | 分子名称: | 4'-PHOSPHOPANTETHEINYL TRANSFERASE SFP, COENZYME A, MAGNESIUM ION | | 著者 | Reuter, K, Mofid, R.M, Marahiel, A.M, Ficner, R. | | 登録日 | 1999-06-17 | | 公開日 | 1999-12-10 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Crystal structure of the surfactin synthetase-activating enzyme sfp: a prototype of the 4'-phosphopantetheinyl transferase superfamily.
EMBO J., 18, 1999
|
|
