7P2P
 
 | | Human Signal Peptidase Complex Paralog A (SPC-A) | | 分子名称: | Signal peptidase complex catalytic subunit SEC11A, Signal peptidase complex subunit 1, Signal peptidase complex subunit 2, ... | | 著者 | Liaci, A.M, Foerster, F. | | 登録日 | 2021-07-06 | | 公開日 | 2021-10-06 | | 最終更新日 | 2022-12-21 | | 実験手法 | ELECTRON MICROSCOPY (4.9 Å) | | 主引用文献 | Structure of the human signal peptidase complex reveals the determinants for signal peptide cleavage.
Mol.Cell, 81, 2021
|
|
6OXQ
 
 | | HIV-1 Protease NL4-3 WT in Complex with UMass8 | | 分子名称: | (3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl {(1S,2R)-1-benzyl-3-[(2-ethylbutyl){[4-(hydroxymethyl)phenyl]sulfonyl}amino]-2-hydroxypropyl}carbamate, Protease NL4-3, SULFATE ION | | 著者 | Lockbaum, G.J, Rusere, L.N, Lee, S.K, Henes, M, Kosovrasti, K, Spielvogel, E, Nalivaika, E.A, Swanstrom, R, KurtYilmaz, N, Schiffer, C.A, Ali, A. | | 登録日 | 2019-05-14 | | 公開日 | 2019-08-21 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.89 Å) | | 主引用文献 | HIV-1 Protease Inhibitors Incorporating Stereochemically Defined P2' Ligands To Optimize Hydrogen Bonding in the Substrate Envelope.
J.Med.Chem., 62, 2019
|
|
6OXV
 
 | | HIV-1 Protease NL4-3 WT in Complex with LR-85 | | 分子名称: | (3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl {(2S,3R)-4-[(2-ethylbutyl)({4-[(1S)-1-hydroxyethyl]phenyl}sulfonyl)amino]-3-hydroxy-1-phenylbutan-2-yl}carbamate, Protease NL4-3 | | 著者 | Lockbaum, G.J, Rusere, L.N, Lee, S.K, Henes, M, Kosovrasti, K, Spielvogel, E, Nalivaika, E.A, Swanstrom, R, KurtYilmaz, N, Schiffer, C.A, Ali, A. | | 登録日 | 2019-05-14 | | 公開日 | 2019-08-21 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.991 Å) | | 主引用文献 | HIV-1 Protease Inhibitors Incorporating Stereochemically Defined P2' Ligands To Optimize Hydrogen Bonding in the Substrate Envelope.
J.Med.Chem., 62, 2019
|
|
6OXY
 
 | | HIV-1 Protease NL4-3 WT in Complex with LR2-19 | | 分子名称: | (3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl {(2S,3R)-4-[({4-[(1S)-1,2-dihydroxyethyl]phenyl}sulfonyl)(2-methylpropyl)amino]-3-hydroxy-1-phenylbutan-2-yl}carbamate, Protease NL4-3, SULFATE ION | | 著者 | Lockbaum, G.J, Rusere, L.N, Lee, S.K, Henes, M, Kosovrasti, K, Spielvogel, E, Nalivaika, E.A, Swanstrom, R, KurtYilmaz, N, Schiffer, C.A, Ali, A. | | 登録日 | 2019-05-14 | | 公開日 | 2019-08-21 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.96 Å) | | 主引用文献 | HIV-1 Protease Inhibitors Incorporating Stereochemically Defined P2' Ligands To Optimize Hydrogen Bonding in the Substrate Envelope.
J.Med.Chem., 62, 2019
|
|
1FUQ
 
 | |
7P11
 
 | | Galectin-8 N-terminal carbohydrate recognition domain in complex with quinoline D-galactal ligand | | 分子名称: | 2-[[(2~{R},3~{R},4~{R})-2-(hydroxymethyl)-3-oxidanyl-3,4-dihydro-2~{H}-pyran-4-yl]oxymethyl]quinoline-7-carboxylic acid, CHLORIDE ION, Galectin-8, ... | | 著者 | Hassan, M, Hakansson, M, Nilsson, J.U, Kovacic, R. | | 登録日 | 2021-07-01 | | 公開日 | 2021-12-15 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Structure-Guided Design of d-Galactal Derivatives with High Affinity and Selectivity for the Galectin-8 N-Terminal Domain.
Acs Med.Chem.Lett., 12, 2021
|
|
7P1M
 
 | | Galectin-8 N-terminal carbohydrate recognition domain in complex with benzimidazole D-galactal ligand | | 分子名称: | 2-[[(2R,3R,4R)-2-(hydroxymethyl)-3-oxidanyl-3,4-dihydro-2H-pyran-4-yl]oxymethyl]-3-methyl-benzimidazole-5-carboxylic acid, CHLORIDE ION, Galectin-8 | | 著者 | Hassan, M, Hakansson, M, Nilsson, J.U, Kovacic, R. | | 登録日 | 2021-07-02 | | 公開日 | 2021-12-15 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.52 Å) | | 主引用文献 | Structure-Guided Design of d-Galactal Derivatives with High Affinity and Selectivity for the Galectin-8 N-Terminal Domain.
Acs Med.Chem.Lett., 12, 2021
|
|
7OVP
 
 | |
6OXT
 
 | | HIV-1 Protease NL4-3 WT in Complex with LR-84 | | 分子名称: | (3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl [(2S,3R)-3-hydroxy-4-{({4-[(1S)-1-hydroxyethyl]phenyl}sulfonyl)[(2S)-2-methylbutyl]amino}-1-phenylbutan-2-yl]carbamate, Protease NL4-3, SULFATE ION | | 著者 | Lockbaum, G.J, Rusere, L.N, Lee, S.K, Henes, M, Kosovrasti, K, Spielvogel, E, Nalivaika, E.A, Swanstrom, R, KurtYilmaz, N, Schiffer, C.A, Ali, A. | | 登録日 | 2019-05-14 | | 公開日 | 2019-08-21 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.861 Å) | | 主引用文献 | HIV-1 Protease Inhibitors Incorporating Stereochemically Defined P2' Ligands To Optimize Hydrogen Bonding in the Substrate Envelope.
J.Med.Chem., 62, 2019
|
|
6OY0
 
 | | HIV-1 Protease NL4-3 WT in Complex with LR2-21 | | 分子名称: | (3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl [(2S,3R)-4-{({4-[(1S)-1,2-dihydroxyethyl]phenyl}sulfonyl)[(2S)-2-methylbutyl]amino}-3-hydroxy-1-phenylbutan-2-yl]carbamate, Protease NL4-3, SULFATE ION | | 著者 | Lockbaum, G.J, Rusere, L.N, Lee, S.K, Henes, M, Kosovrasti, K, Spielvogel, E, Nalivaika, E.A, Swanstrom, R, KurtYilmaz, N, Schiffer, C.A, Ali, A. | | 登録日 | 2019-05-14 | | 公開日 | 2019-08-21 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | HIV-1 Protease Inhibitors Incorporating Stereochemically Defined P2' Ligands To Optimize Hydrogen Bonding in the Substrate Envelope.
J.Med.Chem., 62, 2019
|
|
1GSU
 
 | | AN AVIAN CLASS-MU GLUTATHIONE S-TRANSFERASE, CGSTM1-1 AT 1.94 ANGSTROM RESOLUTION | | 分子名称: | CLASS-MU GLUTATHIONE S-TRANSFERASE, S-HEXYLGLUTATHIONE | | 著者 | Sun, Y.-J, Kuan, C, Tam, M.F, Hsiao, C.-D. | | 登録日 | 1997-09-02 | | 公開日 | 1998-03-04 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.94 Å) | | 主引用文献 | The three-dimensional structure of an avian class-mu glutathione S-transferase, cGSTM1-1 at 1.94 A resolution.
J.Mol.Biol., 278, 1998
|
|
1H6V
 
 | | Mammalian thioredoxin reductase | | 分子名称: | FLAVIN-ADENINE DINUCLEOTIDE, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, THIOREDOXIN REDUCTASE | | 著者 | Sandalova, T, Zhong, L, Lindqvist, Y, Holmgren, A, Schneider, G. | | 登録日 | 2001-06-27 | | 公開日 | 2001-08-14 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Three-Dimensional Structure of a Mammalian Thioredoxin Reductase: Implication for Mechanism and Evolution of a Selenocysteine Dependent Enzyme
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
1HIZ
 
 | | Xylanase T6 (Xt6) from Bacillus Stearothermophilus | | 分子名称: | ENDO-1,4-BETA-XYLANASE, SULFATE ION, alpha-D-galactopyranose, ... | | 著者 | Sainz, G, Tepplitsky, A, Stojanoff, V, Thompson, A, Shoham, Y, Shoham, G. | | 登録日 | 2001-01-05 | | 公開日 | 2002-01-04 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Structure Determination of the Extracellular Xylanase from Geobacillus Stearothermophilus by Selenomethionyl MAD Phasing
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1ESG
 
 | |
1EED
 
 | | X-ray crystallographic analysis of inhibition of endothiapepsin by cyclohexyl renin inhibitors | | 分子名称: | (2S)-2-[[(3S,4S)-5-cyclohexyl-4-[[(4S,5S)-5-[(2-methylpropan-2-yl)oxycarbonylamino]-4-oxidanyl-6-phenyl-hexanoyl]amino]-3-oxidanyl-pentanoyl]amino]-4-methyl-pentanoic acid, ENDOTHIAPEPSIN | | 著者 | Blundell, T.L, Frazao, C, Cooper, J.B. | | 登録日 | 1992-06-15 | | 公開日 | 1994-01-31 | | 最終更新日 | 2017-11-29 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | X-ray crystallographic analysis of inhibition of endothiapepsin by cyclohexyl renin inhibitors.
Biochemistry, 31, 1992
|
|
8P8G
 
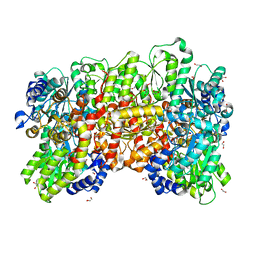 | | Nitrogenase MoFe protein from A. vinelandii beta double mutant D353G/D357G | | 分子名称: | 1,2-ETHANEDIOL, 1,4-DIETHYLENE DIOXIDE, 3-HYDROXY-3-CARBOXY-ADIPIC ACID, ... | | 著者 | Maslac, N, Wagner, T. | | 登録日 | 2023-06-01 | | 公開日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | The Mononuclear Metal-Binding Site of Mo-Nitrogenase Is Not Required for Activity.
Jacs Au, 3, 2023
|
|
4KPR
 
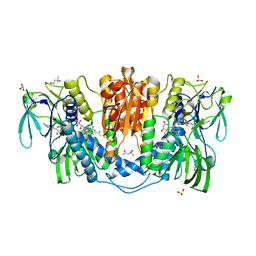 | | Tetrameric form of rat selenoprotein thioredoxin reductase 1 | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, FLAVIN-ADENINE DINUCLEOTIDE, SULFITE ION, ... | | 著者 | Lindqvist, Y, Sandalova, T, Xu, J, Arner, E. | | 登録日 | 2013-05-14 | | 公開日 | 2014-05-14 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | The Trp114 residue of thioredoxin reductase 1 is an electron relay sensor for oxidative stress
To be Published, 2013
|
|
1E94
 
 | | HslV-HslU from E.coli | | 分子名称: | HEAT SHOCK PROTEIN HSLU, HEAT SHOCK PROTEIN HSLV, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | 著者 | Song, H.K, Hartmann, C, Ravishankar, R, Bochtler, M. | | 登録日 | 2000-10-07 | | 公開日 | 2000-11-17 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Mutational Studies on Hslu and its Docking Mode with Hslv
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
1EQW
 
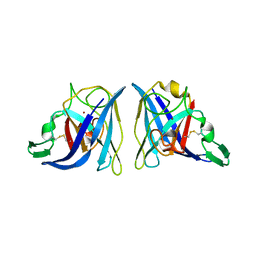 | | CRYSTAL STRUCTURE OF SALMONELLA TYPHIMURIUM CU,ZN SUPEROXIDE DISMUTASE | | 分子名称: | COPPER (II) ION, CU,ZN SUPEROXIDE DISMUTASE, ZINC ION | | 著者 | Pesce, A, Battistoni, A, Stroppolo, M.E, Polizio, F, Nardini, M, Kroll, J.S, Langford, P.R, O'Neill, P, Sette, M, Desideri, A, Bolognesi, M. | | 登録日 | 2000-04-06 | | 公開日 | 2000-09-08 | | 最終更新日 | 2017-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Functional and crystallographic characterization of Salmonella typhimurium Cu,Zn superoxide dismutase coded by the sodCI virulence gene.
J.Mol.Biol., 302, 2000
|
|
8PPS
 
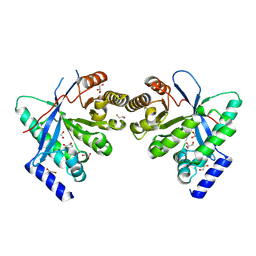 | | Dimeric RbdA EAL, in apo state | | 分子名称: | 1,2-ETHANEDIOL, BETA-MERCAPTOETHANOL, EAL domain-containing protein, ... | | 著者 | Cordery, C.R, Maly, M, Walsh, M.A, Tews, I. | | 登録日 | 2023-07-08 | | 公開日 | 2024-05-15 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Phosphodiesterase activation in the biofilm dispersal protein RbdA and relationship to the biofilm formation protein PA2072 of similar architecture
To Be Published
|
|
8PVL
 
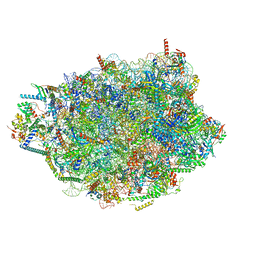 | | Chaetomium thermophilum pre-60S State 7 - pre-5S rotation lacking Utp30/ITS2 - composite structure | | 分子名称: | 26S rRNA, 5.8S rRNA, 5S rRNA, ... | | 著者 | Thoms, M, Cheng, J, Denk, T, Berninghausen, O, Beckmann, R. | | 登録日 | 2023-07-17 | | 公開日 | 2023-12-06 | | 最終更新日 | 2023-12-20 | | 実験手法 | ELECTRON MICROSCOPY (2.19 Å) | | 主引用文献 | Structural insights into coordinating 5S RNP rotation with ITS2 pre-RNA processing during ribosome formation.
Embo Rep., 24, 2023
|
|
8PVK
 
 | | Chaetomium thermophilum pre-60S State 5 - pre-5S rotation - L1 inward - composite structure | | 分子名称: | 26S rRNA, 5.8S rRNA, 5S rRNA, ... | | 著者 | Thoms, M, Cheng, J, Denk, T, Berninghausen, O, Beckmann, R. | | 登録日 | 2023-07-17 | | 公開日 | 2023-12-06 | | 最終更新日 | 2023-12-20 | | 実験手法 | ELECTRON MICROSCOPY (2.55 Å) | | 主引用文献 | Structural insights into coordinating 5S RNP rotation with ITS2 pre-RNA processing during ribosome formation.
Embo Rep., 24, 2023
|
|
8PV8
 
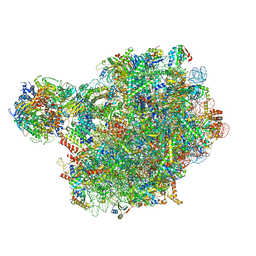 | | Chaetomium thermophilum pre-60S State 4 - post-5S rotation with Rix1 complex without Foot - composite structure | | 分子名称: | 26S rRNA, 5.8S rRNA, 5S rRNA, ... | | 著者 | Thoms, M, Cheng, J, Denk, T, Berninghausen, O, Beckmann, R. | | 登録日 | 2023-07-17 | | 公開日 | 2024-01-10 | | 最終更新日 | 2024-01-17 | | 実験手法 | ELECTRON MICROSCOPY (2.91 Å) | | 主引用文献 | Structural insights into coordinating 5S RNP rotation with ITS2 pre-RNA processing during ribosome formation.
Embo Rep., 24, 2023
|
|
8PO2
 
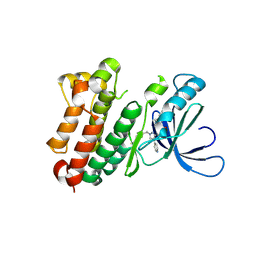 | |
8PNZ
 
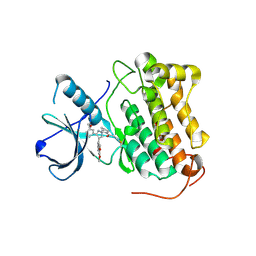 | |
