3DGJ
 
 | | NNFGAIL segment from Islet Amyloid Polypeptide (IAPP or amylin) | | 分子名称: | NNFGAIL peptide | | 著者 | Wiltzius, J.J, Sievers, S.A, Sawaya, M.R, Cascio, D, Eisenberg, D. | | 登録日 | 2008-06-13 | | 公開日 | 2008-07-01 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Atomic structure of the cross-beta spine of islet amyloid polypeptide (amylin).
Protein Sci., 17, 2008
|
|
2XPZ
 
 | | Structure of native yeast LTA4 hydrolase | | 分子名称: | (R,R)-2,3-BUTANEDIOL, LEUKOTRIENE A-4 HYDROLASE, POLYETHYLENE GLYCOL (N=34), ... | | 著者 | Helgstrand, C, Hasan, M, Usyal, H, Haeggstrom, J.Z, Thunnissen, M.M.G.M. | | 登録日 | 2010-08-31 | | 公開日 | 2010-12-29 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | A Leukotriene A(4) Hydrolase-Related Aminopeptidase from Yeast Undergoes Induced Fit Upon Inhibitor Binding.
J.Mol.Biol., 406, 2011
|
|
7X3Y
 
 | | Cryo-EM structure of Coxsackievirus B1 empty particle in complex with nAb 9A3 (CVB1-E:9A3) | | 分子名称: | 9A3 heavy chain, 9A3 light chain, VP2, ... | | 著者 | Zheng, Q, Zhu, R, Sun, H, Cheng, T, Li, S, Xia, N. | | 登録日 | 2022-03-01 | | 公開日 | 2023-06-07 | | 最終更新日 | 2024-10-16 | | 実験手法 | ELECTRON MICROSCOPY (3.32 Å) | | 主引用文献 | Structural basis for the synergistic neutralization of coxsackievirus B1 by a triple-antibody cocktail.
Cell Host Microbe, 30, 2022
|
|
5MDO
 
 | |
7OMT
 
 | | Crystal structure of ProMacrobody 21 with bound maltose | | 分子名称: | HEXAETHYLENE GLYCOL, MAGNESIUM ION, ProMacrobody 21, ... | | 著者 | Botte, M, Ni, D, Schenck, S, Zimmermann, I, Chami, M, Bocquet, N, Egloff, P, Bucher, D, Trabuco, M, Cheng, R.K.Y, Brunner, J.D, Seeger, M.A, Stahlberg, H, Hennig, M. | | 登録日 | 2021-05-24 | | 公開日 | 2022-05-04 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Cryo-EM structures of a LptDE transporter in complex with Pro-macrobodies offer insight into lipopolysaccharide translocation.
Nat Commun, 13, 2022
|
|
7OMM
 
 | | Cryo-EM structure of N. gonorhoeae LptDE in complex with ProMacrobodies (MBPs have not been built de novo) | | 分子名称: | LPS-assembly lipoprotein LptE, LPS-assembly protein LptD, ProMacrobody 21,Maltodextrin-binding protein, ... | | 著者 | Botte, M, Ni, D, Schenck, S, Zimmermann, I, Chami, M, Bocquet, N, Egloff, P, Bucher, D, Trabuco, M, Cheng, R.K.Y, Brunner, J.D, Seeger, M.A, Stahlberg, H, Hennig, M. | | 登録日 | 2021-05-24 | | 公開日 | 2022-05-04 | | 最終更新日 | 2022-05-11 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Cryo-EM structures of a LptDE transporter in complex with Pro-macrobodies offer insight into lipopolysaccharide translocation.
Nat Commun, 13, 2022
|
|
7X35
 
 | | Cryo-EM structure of Coxsackievirus B1 A-particle in complex with nAb 8A10 (CVB1-A:8A10) | | 分子名称: | 8A10 heavy chain, 8A10 light chain, VP2, ... | | 著者 | Zheng, Q, Zhu, R, Sun, H, Cheng, T, Li, S, Xia, N. | | 登録日 | 2022-02-28 | | 公開日 | 2024-07-24 | | 実験手法 | ELECTRON MICROSCOPY (3.19 Å) | | 主引用文献 | Structural basis for the synergistic neutralization of coxsackievirus B1 by a triple-antibody cocktail.
Cell Host Microbe, 30, 2022
|
|
2WKK
 
 | | Identification of the glycan target of the nematotoxic fungal galectin CGL2 in Caenorhabditis elegans | | 分子名称: | GALECTIN-2, GLYCEROL, MAGNESIUM ION, ... | | 著者 | Butschi, A, Titz, A, Waelti, M, Olieric, V, Paschinger, K, Xiaoqiang, G, Seeberger, P.H, Wilson, I.B.H, Aebi, M, Hengartner, M.O, Kuenzler, M. | | 登録日 | 2009-06-14 | | 公開日 | 2010-01-19 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Caenorhabditis Elegans N-Glycan Core Beta-Galactoside Confers Sensitivity Towards Nematotoxic Fungal Galectin Cgl2.
Plos Pathog., 6, 2010
|
|
3JQY
 
 | | Crystal Structure of the polySia specific acetyltransferase NeuO | | 分子名称: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, Polysialic acid O-acetyltransferase | | 著者 | Schulz, E.-C, Bergfeld, A, Muehlenhoff, M, Ficner, R. | | 登録日 | 2009-09-08 | | 公開日 | 2010-08-18 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.699 Å) | | 主引用文献 | Crystal structure analysis of the polysialic acid specific O-acetyltransferase NeuO
PLoS ONE, 6, 2011
|
|
5WL8
 
 | | Crystal structure of chalcone isomerase engineered from ancestral inference (epR4) | | 分子名称: | Engineered Chalcone Isomerase epR4, FORMIC ACID | | 著者 | Burke, J.R, Kaltenbach, M, Tawfik, D.S, Noel, J.P. | | 登録日 | 2017-07-25 | | 公開日 | 2018-05-09 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.58 Å) | | 主引用文献 | Evolution of chalcone isomerase from a noncatalytic ancestor.
Nat. Chem. Biol., 14, 2018
|
|
2HHT
 
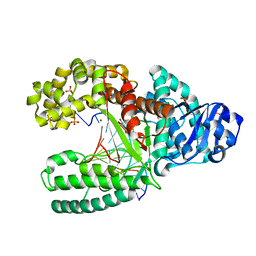 | | C:O6-methyl-guanine pair in the polymerase-2 basepair position | | 分子名称: | 5'-D(*GP*CP*GP*AP*TP*CP*AP*GP*CP*TP*CP*G)-3', 5'-D(*GP*TP*AP*CP*(6OG)P*AP*GP*CP*TP*GP*AP*TP*CP*GP*CP*A)-3', DNA polymerase I, ... | | 著者 | Warren, J.J, Forsberg, L.J, Beese, L.S. | | 登録日 | 2006-06-28 | | 公開日 | 2006-12-12 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.05 Å) | | 主引用文献 | The structural basis for the mutagenicity of O6-methyl-guanine lesions.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
2X0W
 
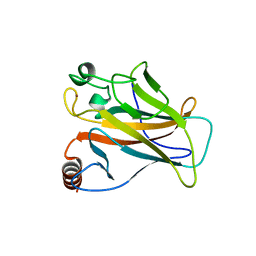 | | STRUCTURE OF THE P53 CORE DOMAIN MUTANT Y220C BOUND TO 5,6-dimethoxy- 2-methylbenzothiazole | | 分子名称: | 5,6-DIMETHOXY-2-METHYL-1,3-BENZOTHIAZOLE, CELLULAR TUMOR ANTIGEN P53, ZINC ION | | 著者 | Kaar, J.L, Basse, N, Joerger, A.C, Fersht, A.R. | | 登録日 | 2009-12-17 | | 公開日 | 2010-01-26 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Toward the Rational Design of P53-Stabilizing Drugs: Probing the Surface of the Oncogenic Y220C Mutant.
Chem.Biol., 17, 2010
|
|
5WKR
 
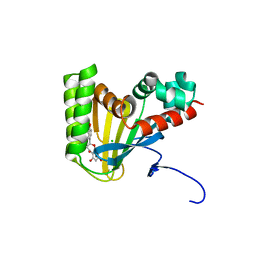 | | Crystal structure of chalcone isomerase engineered from ancestral inference complexed with naringenin (ancCC) | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, CHLORIDE ION, Engineered Chalcone Isomerase ancCC, ... | | 著者 | Burke, J.R, Kaltenbach, M, Tawfik, D.S, Noel, J.P. | | 登録日 | 2017-07-25 | | 公開日 | 2018-05-09 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Evolution of chalcone isomerase from a noncatalytic ancestor.
Nat. Chem. Biol., 14, 2018
|
|
5MDQ
 
 | |
5WL4
 
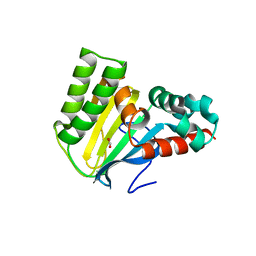 | | Crystal structure of chalcone isomerase engineered from ancestral inference (ancR3) | | 分子名称: | Engineered Chalcone Isomerase ancR3, FORMIC ACID | | 著者 | Burke, J.R, Kaltenbach, M, Tawfik, D.S, Noel, J.P. | | 登録日 | 2017-07-25 | | 公開日 | 2018-05-09 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Evolution of chalcone isomerase from a noncatalytic ancestor.
Nat. Chem. Biol., 14, 2018
|
|
5MXN
 
 | | Atomic model of the VipA/VipB/Hcp, the type six secretion system non-contractile sheath-tube of Vibrio cholerae from cryo-EM | | 分子名称: | Haemolysin co-regulated protein, Type VI secretion protein | | 著者 | Wang, J, Brackmann, M, Castano-Diez, D, Kudryashev, M, Goldie, K, Maier, T, Stahlberg, H, Basler, M. | | 登録日 | 2017-01-23 | | 公開日 | 2017-08-02 | | 最終更新日 | 2024-05-08 | | 実験手法 | ELECTRON MICROSCOPY (3.7 Å) | | 主引用文献 | Cryo-EM structure of the extended type VI secretion system sheath-tube complex.
Nat Microbiol, 2, 2017
|
|
7OS7
 
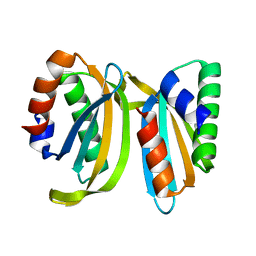 | | Circular permutant of ribosomal protein S6, swap helix 2, L75A, A92K mutant | | 分子名称: | 30S ribosomal protein S6,30S ribosomal protein S6 | | 著者 | Wang, H, Logan, D.T, Oliveberg, M. | | 登録日 | 2021-06-08 | | 公開日 | 2022-06-22 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | Circular permutant of ribosomal protein S6, swap helix 2, L75A, A92K mutant
To Be Published
|
|
3GD6
 
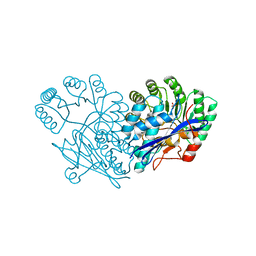 | | Crystal structure of divergent enolase from Oceanobacillus iheyensis complexed with phosphate | | 分子名称: | Muconate cycloisomerase, PHOSPHATE ION | | 著者 | Fedorov, A.A, Fedorov, E.V, Sauder, J.M, Burley, S.K, Gerlt, J.A, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | 登録日 | 2009-02-23 | | 公開日 | 2009-03-03 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Crystal structure of divergent enolase from Oceanobacillus iheyensis complexed with phosphate.
To be Published
|
|
5WKS
 
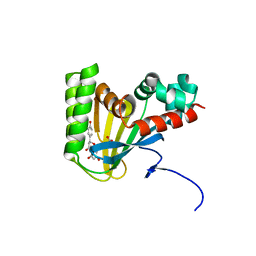 | | Crystal structure of chalcone isomerase engineered from ancestral inference complexed with naringenin (ancR1) | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, Engineered Chalcone Isomerase ancR1, FORMIC ACID, ... | | 著者 | Burke, J.R, Kaltenbach, M, Tawfik, D.S, Noel, J.P. | | 登録日 | 2017-07-25 | | 公開日 | 2018-05-09 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Evolution of chalcone isomerase from a noncatalytic ancestor.
Nat. Chem. Biol., 14, 2018
|
|
7X38
 
 | | Cryo-EM structure of Coxsackievirus B1 empty particle in complex with nAb 8A10 (CVB1-E:8A10) | | 分子名称: | 8A10 heavy chain, 8A10 light chain, VP2, ... | | 著者 | Zheng, Q, Zhu, R, Sun, H, Cheng, T, Li, S, Xia, N. | | 登録日 | 2022-02-28 | | 公開日 | 2022-09-28 | | 実験手法 | ELECTRON MICROSCOPY (3.52 Å) | | 主引用文献 | Structural basis for the synergistic neutralization of coxsackievirus B1 by a triple-antibody cocktail.
Cell Host Microbe, 30, 2022
|
|
7X3F
 
 | | Cryo-EM structure of Coxsackievirus B1 A-particle in complex with nAb 9A3 (CVB1-A:9A3) | | 分子名称: | 9A3 heavy chain, 9A3 light chain, VP2, ... | | 著者 | Zheng, Q, Zhu, R, Sun, H, Cheng, T, Li, S, Xia, N. | | 登録日 | 2022-02-28 | | 公開日 | 2022-09-28 | | 実験手法 | ELECTRON MICROSCOPY (3.52 Å) | | 主引用文献 | Structural basis for the synergistic neutralization of coxsackievirus B1 by a triple-antibody cocktail.
Cell Host Microbe, 30, 2022
|
|
5WL5
 
 | | Crystal structure of chalcone isomerase engineered from ancestral inference (ancR5) | | 分子名称: | CHLORIDE ION, Engineered Chalcone Isomerase ancR5, SULFATE ION | | 著者 | Burke, J.R, Kaltenbach, M, Tawfik, D.S, Noel, J.P. | | 登録日 | 2017-07-25 | | 公開日 | 2018-05-09 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.513 Å) | | 主引用文献 | Evolution of chalcone isomerase from a noncatalytic ancestor.
Nat. Chem. Biol., 14, 2018
|
|
5WL7
 
 | | Crystal structure of chalcone isomerase engineered from ancestral inference (ancCHI*) | | 分子名称: | CHLORIDE ION, Engineered Chalcone Isomerase ancCHI* | | 著者 | Burke, J.R, Kaltenbach, M, Tawfik, D.S, Noel, J.P. | | 登録日 | 2017-07-25 | | 公開日 | 2018-05-09 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Evolution of chalcone isomerase from a noncatalytic ancestor.
Nat. Chem. Biol., 14, 2018
|
|
7X2G
 
 | | Cryo-EM structure of Coxsackievirus B1 empty particle in complex with nAb nAb 2E6 (CVB1-E:2E6) | | 分子名称: | 2E6 heavy chain, 2E6 light chain, VP2, ... | | 著者 | Zheng, Q, Zhu, R, Sun, H, Cheng, T, Li, S, Xia, N. | | 登録日 | 2022-02-25 | | 公開日 | 2022-09-28 | | 最終更新日 | 2022-10-05 | | 実験手法 | ELECTRON MICROSCOPY (3.58 Å) | | 主引用文献 | Structural basis for the synergistic neutralization of coxsackievirus B1 by a triple-antibody cocktail.
Cell Host Microbe, 30, 2022
|
|
7X2O
 
 | | Cryo-EM structure of Coxsackievirus B1 mature virion in complex with nAb 2E6 (CVB1-M:2E6) | | 分子名称: | 2E6 heavy chain, 2E6 light chain, Capsid protein VP4, ... | | 著者 | Zheng, Q, Zhu, R, Sun, H, Cheng, T, Li, S, Xia, N. | | 登録日 | 2022-02-25 | | 公開日 | 2022-09-28 | | 最終更新日 | 2022-10-05 | | 実験手法 | ELECTRON MICROSCOPY (3.15 Å) | | 主引用文献 | Structural basis for the synergistic neutralization of coxsackievirus B1 by a triple-antibody cocktail.
Cell Host Microbe, 30, 2022
|
|
