2FAX
 
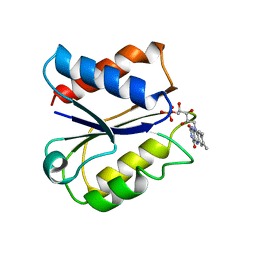 | | CLOSTRIDIUM BEIJERINCKII FLAVODOXIN MUTANT: N137A OXIDIZED (150K) | | 分子名称: | FLAVIN MONONUCLEOTIDE, FLAVODOXIN | | 著者 | Ludwig, M.L, Pattridge, K.A, Metzger, A.L, Dixon, M.M, Eren, M, Feng, Y, Swenson, R. | | 登録日 | 1996-12-24 | | 公開日 | 1997-03-12 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Control of oxidation-reduction potentials in flavodoxin from Clostridium beijerinckii: the role of conformation changes.
Biochemistry, 36, 1997
|
|
1LDE
 
 | |
1MYN
 
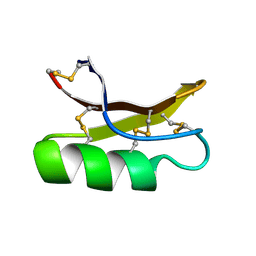 | | SOLUTION STRUCTURE OF DROSOMYCIN, THE FIRST INDUCIBLE ANTIFUNGAL PROTEIN FROM INSECTS, NMR, 15 STRUCTURES | | 分子名称: | DROSOMYCIN | | 著者 | Landon, C, Sodano, P, Hetru, C, Hoffmann, J.A, Ptak, M. | | 登録日 | 1996-12-26 | | 公開日 | 1997-12-31 | | 最終更新日 | 2022-02-23 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of drosomycin, the first inducible antifungal protein from insects.
Protein Sci., 6, 1997
|
|
1UXD
 
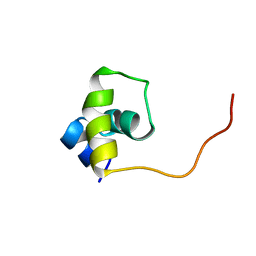 | | Fructose repressor DNA-binding domain, NMR, 34 structures | | 分子名称: | FRUCTOSE REPRESSOR | | 著者 | Penin, F, Geourjon, C, Montserret, R, Bockmann, A, Lesage, A, Yang, Y, Bonod-Bidaud, C, Cortay, J.C, Negre, D, Cozzone, A.J, Deleage, G. | | 登録日 | 1996-12-26 | | 公開日 | 1997-04-01 | | 最終更新日 | 2024-05-01 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Three-dimensional structure of the DNA-binding domain of the fructose repressor from Escherichia coli by 1H and 15N NMR.
J.Mol.Biol., 270, 1997
|
|
1UXC
 
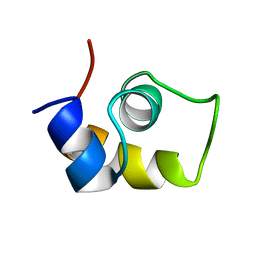 | | FRUCTOSE REPRESSOR DNA-BINDING DOMAIN, NMR, MINIMIZED STRUCTURE | | 分子名称: | FRUCTOSE REPRESSOR | | 著者 | Penin, F, Geourjon, C, Montserret, R, Bockmann, A, Lesage, A, Yang, Y, Bonod-Bidaud, C, Cortay, J.C, Negre, D, Cozzone, A.J, Deleage, G. | | 登録日 | 1996-12-26 | | 公開日 | 1997-04-21 | | 最終更新日 | 2024-05-01 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Three-dimensional structure of the DNA-binding domain of the fructose repressor from Escherichia coli by 1H and 15N NMR.
J.Mol.Biol., 270, 1997
|
|
1MF2
 
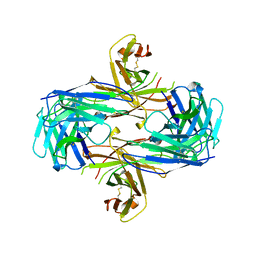 | | ANTI HIV1 PROTEASE FAB COMPLEX | | 分子名称: | MONOCLONAL ANTIBODY F11.2.32 | | 著者 | Lescar, J, Bentley, G.A. | | 登録日 | 1996-12-27 | | 公開日 | 1997-12-31 | | 最終更新日 | 2023-08-09 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Three-dimensional structure of an Fab-peptide complex: structural basis of HIV-1 protease inhibition by a monoclonal antibody.
J.Mol.Biol., 267, 1997
|
|
2HRP
 
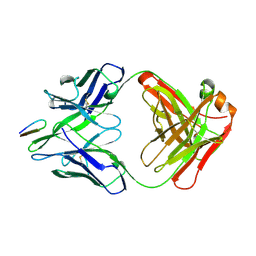 | | ANTIGEN-ANTIBODY COMPLEX | | 分子名称: | HIV-1 PROTEASE PEPTIDE, MONOCLONAL ANTIBODY F11.2.32 | | 著者 | Lescar, J, Bentley, G.A. | | 登録日 | 1996-12-27 | | 公開日 | 1997-12-31 | | 最終更新日 | 2023-08-09 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Three-dimensional structure of an Fab-peptide complex: structural basis of HIV-1 protease inhibition by a monoclonal antibody.
J.Mol.Biol., 267, 1997
|
|
1FFH
 
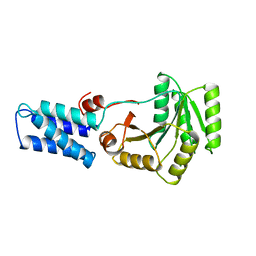 | | N AND GTPASE DOMAINS OF THE SIGNAL SEQUENCE RECOGNITION PROTEIN FFH FROM THERMUS AQUATICUS | | 分子名称: | FFH, MAGNESIUM ION | | 著者 | Freymann, D.M, Keenan, R.J, Stroud, R.M, Walter, P. | | 登録日 | 1996-12-30 | | 公開日 | 1997-12-31 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.05 Å) | | 主引用文献 | Structure of the conserved GTPase domain of the signal recognition particle.
Nature, 385, 1997
|
|
1GAB
 
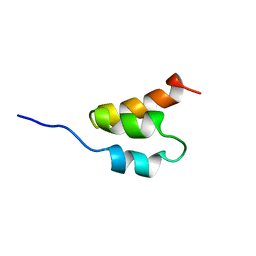 | | STRUCTURE OF AN ALBUMIN-BINDING DOMAIN, NMR, 20 STRUCTURES | | 分子名称: | PROTEIN PAB | | 著者 | Johansson, M.U, De Chateau, M, Wikstrom, M, Forsen, S, Drakenberg, T, Bjorck, L. | | 登録日 | 1996-12-30 | | 公開日 | 1997-07-07 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of the albumin-binding GA module: a versatile bacterial protein domain.
J.Mol.Biol., 266, 1997
|
|
1YUI
 
 | | SOLUTION NMR STRUCTURE OF THE GAGA FACTOR/DNA COMPLEX, REGULARIZED MEAN STRUCTURE | | 分子名称: | DNA (5'-D(*GP*CP*CP*GP*AP*GP*AP*GP*TP*AP*C)-3'), DNA (5'-D(*GP*TP*AP*CP*TP*CP*TP*CP*GP*GP*C)-3'), GAGA-FACTOR, ... | | 著者 | Clore, G.M, Omichinski, J.G, Gronenborn, A.M. | | 登録日 | 1996-12-31 | | 公開日 | 1997-12-31 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | The solution structure of a specific GAGA factor-DNA complex reveals a modular binding mode.
Nat.Struct.Biol., 4, 1997
|
|
1YUJ
 
 | | SOLUTION NMR STRUCTURE OF THE GAGA FACTOR/DNA COMPLEX, 50 STRUCTURES | | 分子名称: | DNA (5'-D(*GP*CP*CP*GP*AP*GP*AP*GP*TP*AP*C)-3'), DNA (5'-D(*GP*TP*AP*CP*TP*CP*TP*CP*GP*GP*C)-3'), GAGA-FACTOR, ... | | 著者 | Clore, G.M, Omichinski, J.G, Gronenborn, A.M. | | 登録日 | 1996-12-31 | | 公開日 | 1997-12-31 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | The solution structure of a specific GAGA factor-DNA complex reveals a modular binding mode.
Nat.Struct.Biol., 4, 1997
|
|
2PIK
 
 | | CALICHEAMICIN GAMMA1I-DNA COMPLEX, NMR, 6 STRUCTURES | | 分子名称: | 2,4-dideoxy-4-(ethylamino)-3-O-methyl-alpha-L-threo-pentopyranose-(1-2)-4-amino-4,6-dideoxy-beta-D-glucopyranose, 2,6-dideoxy-4-thio-beta-D-allopyranose, 3-O-methyl-alpha-L-rhamnopyranose, ... | | 著者 | Kumar, R.A, Ikemoto, N, Patel, D.J. | | 登録日 | 1996-12-31 | | 公開日 | 1997-05-15 | | 最終更新日 | 2024-05-01 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of the calicheamicin gamma 1I-DNA complex.
J.Mol.Biol., 265, 1997
|
|
1XCA
 
 | | APO-CELLULAR RETINOIC ACID BINDING PROTEIN II | | 分子名称: | CELLULAR RETINOIC ACID BINDING PROTEIN TYPE II | | 著者 | Chen, X, Ji, X. | | 登録日 | 1996-12-31 | | 公開日 | 1998-07-01 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Crystal structure of apo-cellular retinoic acid-binding protein type II (R111M) suggests a mechanism of ligand entry.
J.Mol.Biol., 278, 1998
|
|
1EZA
 
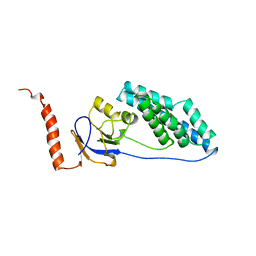 | |
1EZC
 
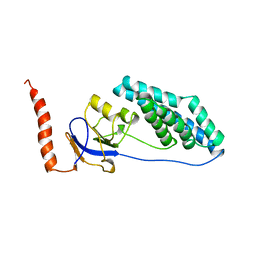 | |
1EZB
 
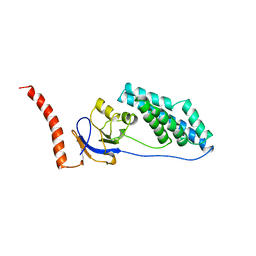 | |
1EZD
 
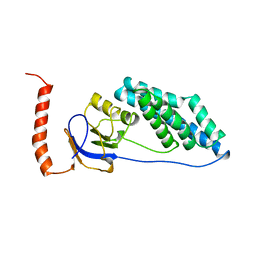 | |
1KPD
 
 | |
305D
 
 | |
2TSY
 
 | | CRYSTAL STRUCTURES OF MUTANT (BETAK87T) TRYPTOPHAN SYNTHASE ALPHA2 BETA2 COMPLEX WITH LIGANDS BOUND TO THE ACTIVE SITES OF THE ALPHA AND BETA SUBUNITS REVEAL LIGAND-INDUCED CONFORMATIONAL CHANGES | | 分子名称: | SN-GLYCEROL-3-PHOSPHATE, SODIUM ION, TRYPTOPHAN SYNTHASE, ... | | 著者 | Rhee, S, Parris, K.D, Hyde, C.C, Ahmed, S.A, Miles, E.W, Davies, D.R. | | 登録日 | 1997-01-03 | | 公開日 | 1997-04-01 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Crystal structures of a mutant (betaK87T) tryptophan synthase alpha2beta2 complex with ligands bound to the active sites of the alpha- and beta-subunits reveal ligand-induced conformational changes.
Biochemistry, 36, 1997
|
|
306D
 
 | |
304D
 
 | |
2TRC
 
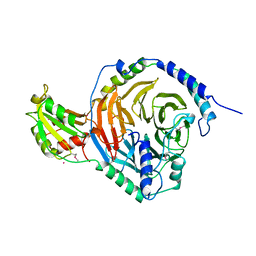 | | PHOSDUCIN/TRANSDUCIN BETA-GAMMA COMPLEX | | 分子名称: | GADOLINIUM ATOM, PHOSDUCIN, TRANSDUCIN | | 著者 | Gaudet, R, Bohm, A, Sigler, P.B. | | 登録日 | 1997-01-06 | | 公開日 | 1997-06-05 | | 最終更新日 | 2019-08-14 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Crystal structure at 2.4 angstroms resolution of the complex of transducin betagamma and its regulator, phosducin.
Cell(Cambridge,Mass.), 87, 1996
|
|
3DBV
 
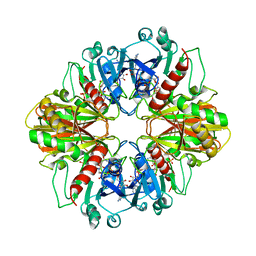 | | GLYCERALDEHYDE-3-PHOSPHATE DEHYDROGENASE MUTANT WITH LEU 33 REPLACED BY THR, THR 34 REPLACED BY GLY, ASP 36 REPLACED BY GLY, LEU 187 REPLACED BY ALA, AND PRO 188 REPLACED BY SER COMPLEXED WITH NAD+ | | 分子名称: | GLYCERALDEHYDE-3-PHOSPHATE DEHYDROGENASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SULFATE ION | | 著者 | Didierjean, C, Rahuel-Clermont, S, Vitoux, B, Dideberg, O, Branlant, G, Aubry, A. | | 登録日 | 1997-01-06 | | 公開日 | 1997-07-07 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.45 Å) | | 主引用文献 | A crystallographic comparison between mutated glyceraldehyde-3-phosphate dehydrogenases from Bacillus stearothermophilus complexed with either NAD+ or NADP+.
J.Mol.Biol., 268, 1997
|
|
4DBV
 
 | | GLYCERALDEHYDE-3-PHOSPHATE DEHYDROGENASE MUTANT WITH LEU 33 REPLACED BY THR, THR 34 REPLACED BY GLY, ASP 36 REPLACED BY GLY, LEU 187 REPLACED BY ALA, AND PRO 188 REPLACED BY SER COMPLEXED WITH NADP+ | | 分子名称: | GLYCERALDEHYDE-3-PHOSPHATE DEHYDROGENASE, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, SULFATE ION | | 著者 | Didierjean, C, Rahuel-Clermont, S, Vitoux, B, Dideberg, O, Branlant, G, Aubry, A. | | 登録日 | 1997-01-06 | | 公開日 | 1997-07-07 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | A crystallographic comparison between mutated glyceraldehyde-3-phosphate dehydrogenases from Bacillus stearothermophilus complexed with either NAD+ or NADP+.
J.Mol.Biol., 268, 1997
|
|
