6KVO
 
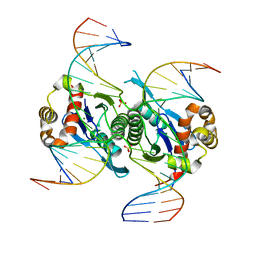 | | Crystal structure of chloroplast resolvase in complex with Holliday junction | | 分子名称: | DNA (5'-D(*AP*CP*AP*AP*CP*AP*GP*AP*TP*GP*AP*TP*GP*GP*AP*GP*CP*T)-3'), DNA (5'-D(*GP*CP*CP*TP*TP*GP*CP*TP*TP*GP*GP*AP*CP*AP*TP*CP*TP*T)-3'), DNA (5'-D(P*AP*AP*GP*AP*TP*GP*TP*CP*CP*AP*TP*CP*TP*GP*TP*TP*GP*T)-3'), ... | | 著者 | Yan, J.J, Hong, S.X, Guan, Z.Y, Yin, P. | | 登録日 | 2019-09-05 | | 公開日 | 2020-04-08 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Structural insights into sequence-dependent Holliday junction resolution by the chloroplast resolvase MOC1.
Nat Commun, 11, 2020
|
|
7M4R
 
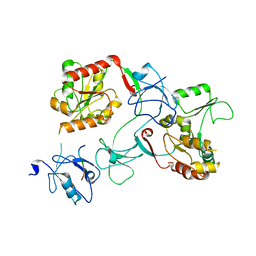 | |
1HJP
 
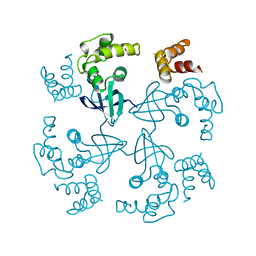 | | HOLLIDAY JUNCTION BINDING PROTEIN RUVA FROM E. COLI | | 分子名称: | RUVA | | 著者 | Nishino, T, Ariyoshi, M, Iwasaki, H, Shinagawa, H, Morikawa, K. | | 登録日 | 1997-08-21 | | 公開日 | 1998-02-25 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Functional Analyses of the Domain Structure in the Holliday Junction Binding Protein Ruva
Structure, 6, 1998
|
|
4EZ2
 
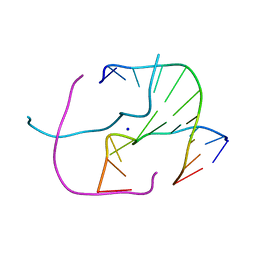 | |
3O2R
 
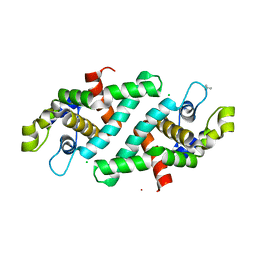 | | Structural flexibility in region involved in dimer formation of nuclease domain of Ribonuclase III (rnc) from Campylobacter jejuni | | 分子名称: | CHLORIDE ION, Ribonuclease III | | 著者 | Minasov, G, Halavaty, A, Shuvalova, L, Dubrovska, I, Winsor, J, Papazisi, L, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2010-07-22 | | 公開日 | 2010-08-04 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.251 Å) | | 主引用文献 | Structural Flexibility in Region Involved in Dimer Formation of Nuclease Domain of Ribonuclase III (rnc) from Campylobacter jejuni.
TO BE PUBLISHED
|
|
3PNU
 
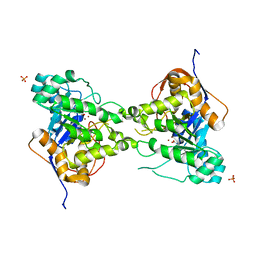 | | 2.4 Angstrom Crystal Structure of Dihydroorotase (pyrC) from Campylobacter jejuni. | | 分子名称: | Dihydroorotase, PHOSPHATE ION, ZINC ION | | 著者 | Minasov, G, Halavaty, A, Shuvalova, L, Dubrovska, I, Winsor, J, Papazisi, L, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2010-11-19 | | 公開日 | 2010-12-01 | | 最終更新日 | 2023-12-06 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | 2.4 Angstrom Crystal Structure of Dihydroorotase (pyrC) from Campylobacter jejuni.
TO BE PUBLISHED
|
|
1FLO
 
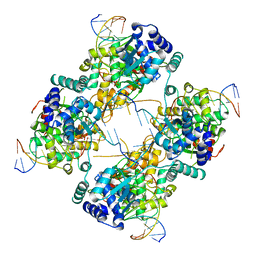 | | FLP Recombinase-Holliday Junction Complex I | | 分子名称: | FLP RECOMBINASE, PHOSPHONIC ACID, SYMMETRIZED FRT DNA SITES | | 著者 | Chen, Y, Narendra, U, Iype, L.E, Cox, M.M, Rice, P.A. | | 登録日 | 2000-08-14 | | 公開日 | 2000-09-04 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.65 Å) | | 主引用文献 | Crystal structure of a Flp recombinase-Holliday junction complex: assembly of an active oligomer by helix swapping.
Mol.Cell, 6, 2000
|
|
7X7P
 
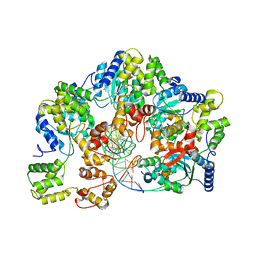 | | CryoEM structure of dsDNA-RuvB-RuvA domain3 complex | | 分子名称: | DNA, Holliday junction ATP-dependent DNA helicase RuvA, Holliday junction ATP-dependent DNA helicase RuvB | | 著者 | Lin, Z, Qu, Q, Zhang, X, Zhou, Z. | | 登録日 | 2022-03-10 | | 公開日 | 2023-03-15 | | 最終更新日 | 2023-09-20 | | 実験手法 | ELECTRON MICROSCOPY (7.02 Å) | | 主引用文献 | Cryo-EM structure of the RuvAB-Holliday junction intermediate complex from Pseudomonas aeruginosa.
Front Plant Sci, 14, 2023
|
|
7XHJ
 
 | |
7X5B
 
 | | Crystal structure of RuvB | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, Holliday junction ATP-dependent DNA helicase RuvB | | 著者 | Lin, Z, Qu, Q, Zhang, X, Zhou, Z, Dai, L. | | 登録日 | 2022-03-04 | | 公開日 | 2023-03-08 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.16 Å) | | 主引用文献 | Cryo-EM structure of the RuvAB-Holliday junction intermediate complex from Pseudomonas aeruginosa.
Front Plant Sci, 14, 2023
|
|
8E9B
 
 | |
5VLG
 
 | | Crystal structure of EilR in complex with malachite green | | 分子名称: | MALACHITE GREEN, Regulatory protein TetR | | 著者 | Pereira, J.H, Ruegg, T.L, Chen, J, Novichkov, P, DeGiovani, A, Tomaleri, G.P, Singer, S, Simmons, B, Thelen, M, Adams, P.D. | | 登録日 | 2017-04-25 | | 公開日 | 2018-06-27 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.932 Å) | | 主引用文献 | Jungle Express is a versatile repressor system for tight transcriptional control.
Nat Commun, 9, 2018
|
|
8EFV
 
 | |
8EWU
 
 | | X-ray structure of the GDP-6-deoxy-4-keto-D-lyxo-heptose-4-reductase from Campylobacter jejuni HS:15 | | 分子名称: | 1,2-ETHANEDIOL, GDP-L-fucose synthase, GUANOSINE-5'-DIPHOSPHATE, ... | | 著者 | Thoden, J.B, Xiang, D.F, Ghosh, M.K, Riegert, A.S, Raushel, F.M, Holden, H.M. | | 登録日 | 2022-10-24 | | 公開日 | 2022-11-09 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.45 Å) | | 主引用文献 | Bifunctional Epimerase/Reductase Enzymes Facilitate the Modulation of 6-Deoxy-Heptoses Found in the Capsular Polysaccharides of Campylobacter jejuni.
Biochemistry, 62, 2023
|
|
8R7Q
 
 | |
8R7P
 
 | |
7SPL
 
 | | [2T3] Self-assembling 3D DNA triangle with three inter-junction base pairs containing the L1 junction and a zero-linked center strand | | 分子名称: | 2'-DEOXYCYTIDINE-5'-MONOPHOSPHATE, DNA (5'-D(*GP*AP*C)-3'), DNA (5'-D(*TP*CP*TP*GP*AP*TP*GP*TP*CP*C)-3'), ... | | 著者 | Vecchioni, S, Lu, B, Sha, R, Ohayon, Y.P, Seeman, N.C. | | 登録日 | 2021-11-02 | | 公開日 | 2022-11-09 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (6.09 Å) | | 主引用文献 | The Rule of Thirds: Controlling Junction Chirality and Polarity in 3D DNA Tiles.
Small, 19, 2023
|
|
8SWD
 
 | |
2G1E
 
 | | Solution Structure of TA0895 | | 分子名称: | hypothetical protein Ta0895 | | 著者 | Yeo, I.Y, Hong, E, Jung, J, Lee, W, Yee, A, Arrowsmith, C.H. | | 登録日 | 2006-02-14 | | 公開日 | 2006-12-26 | | 最終更新日 | 2024-05-29 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of TA0895, a MoaD homologue from Thermoplasma acidophilum
Proteins, 65, 2006
|
|
6LW3
 
 | | Crystal structure of RuvC from Pseudomonas aeruginosa | | 分子名称: | Crossover junction endodeoxyribonuclease RuvC | | 著者 | Hu, Y, He, Y, Lin, Z. | | 登録日 | 2020-02-07 | | 公開日 | 2020-02-26 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.38 Å) | | 主引用文献 | Biochemical and structural characterization of the Holliday junction resolvase RuvC from Pseudomonas aeruginosa.
Biochem.Biophys.Res.Commun., 525, 2020
|
|
1S1L
 
 | |
7AQK
 
 | | Model of the actin filament Arp2/3 complex branch junction in cells | | 分子名称: | Actin, alpha skeletal muscle, ACTA1, ... | | 著者 | Faessler, F, Dimchev, G, Hodirnau, V.V, Wan, W, Schur, F.K.M. | | 登録日 | 2020-10-22 | | 公開日 | 2020-12-02 | | 最終更新日 | 2024-05-01 | | 実験手法 | ELECTRON MICROSCOPY (9 Å) | | 主引用文献 | Cryo-electron tomography structure of Arp2/3 complex in cells reveals new insights into the branch junction.
Nat Commun, 11, 2020
|
|
5TG3
 
 | | Crystal Structure of Dioclea reflexa seed lectin (DrfL) in complex with X-Man | | 分子名称: | 5-bromo-4-chloro-1H-indol-3-yl alpha-D-mannopyranoside, CALCIUM ION, Dioclea reflexa lectin, ... | | 著者 | Santiago, M.Q, Correia, J.L.A, Pinto-Junior, V.R, Osterne, V.J.S, Pereira, R.I, Silva-Filho, J.C, Lossio, C.F, Rocha, B.A.M, Delatorre, P, Neco, A.H.B, Araripe, D.A, Nascimento, K.S, Cavada, B.S. | | 登録日 | 2016-09-27 | | 公開日 | 2017-02-15 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.765 Å) | | 主引用文献 | Structural studies of a vasorelaxant lectin from Dioclea reflexa Hook seeds: Crystal structure, molecular docking and dynamics.
Int. J. Biol. Macromol., 98, 2017
|
|
8D93
 
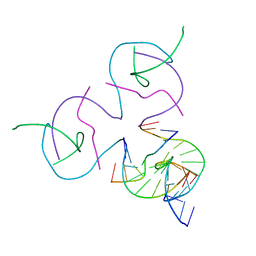 | | [2T7] Self-assembling tensegrity triangle with R3 symmetry at 2.96 A resolution, update and junction cut for entry 3GBI | | 分子名称: | DNA (5'-D(*GP*AP*GP*CP*AP*GP*CP*CP*TP*GP*TP*A)-3'), DNA (5'-D(*TP*CP*TP*GP*AP*TP*GP*TP*GP*GP*CP*TP*GP*C)-3'), DNA (5'-D(P*AP*CP*AP*CP*CP*GP*T)-3'), ... | | 著者 | Vecchioni, S, Woloszyn, K, Lu, B, Sha, R, Ohayon, Y.P, Seeman, N.C. | | 登録日 | 2022-06-09 | | 公開日 | 2023-01-25 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.96 Å) | | 主引用文献 | The Rule of Thirds: Controlling Junction Chirality and Polarity in 3D DNA Tiles.
Small, 19, 2023
|
|
5ER7
 
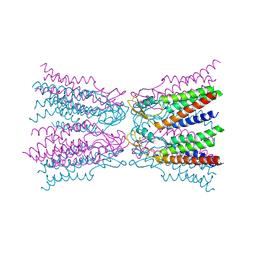 | | Connexin-26 Bound to Calcium | | 分子名称: | CALCIUM ION, Gap junction beta-2 protein | | 著者 | Purdy, M.D, Bennett, B.C, Baker, K.A, Yeager, M.J. | | 登録日 | 2015-11-13 | | 公開日 | 2016-01-27 | | 最終更新日 | 2019-12-04 | | 実験手法 | X-RAY DIFFRACTION (3.286 Å) | | 主引用文献 | An electrostatic mechanism for Ca(2+)-mediated regulation of gap junction channels.
Nat Commun, 7, 2016
|
|
