8XCS
 
 | | Cryo-EM structure of Glutamate dehydrogenase from Thermococcus profundus in complex with NADPH, AKG and NH4 in the initial stage of reaction | | 分子名称: | 2-OXOGLUTARIC ACID, AMMONIUM ION, Glutamate dehydrogenase, ... | | 著者 | Wakabayashi, T, Oide, M, Nakasako, M. | | 登録日 | 2023-12-10 | | 公開日 | 2023-12-27 | | 最終更新日 | 2024-06-19 | | 実験手法 | ELECTRON MICROSCOPY (2.63 Å) | | 主引用文献 | CryoEM-sampling of metastable conformations appearing in cofactor-ligand association and catalysis of glutamate dehydrogenase.
Sci Rep, 14, 2024
|
|
8XCR
 
 | |
8XCQ
 
 | |
8XCP
 
 | |
8XCO
 
 | |
8XCN
 
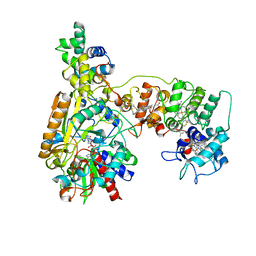 | | Cryo-EM Structure of Membrane-bound Fructose Dehydrogenase from Gluconobacter japonicus variant-N1190A | | 分子名称: | FE3-S4 CLUSTER, FLAVIN-ADENINE DINUCLEOTIDE, Fructose dehydrogenase cytochrome subunit, ... | | 著者 | Fukawa, E, Miyata, T, Makino, F, Adachi, T, Suzuki, Y, Tanaka, H, Namba, K, Sowa, K, Kitazumi, Y, Shirai, O. | | 登録日 | 2023-12-09 | | 公開日 | 2024-05-22 | | 実験手法 | ELECTRON MICROSCOPY (3.02 Å) | | 主引用文献 | Structural and electrochemical elucidation of biocatalytic mechanisms in direct electron transfer-type D-fructose dehydrogenase.
Electrochim Acta, 490, 2024
|
|
8XCM
 
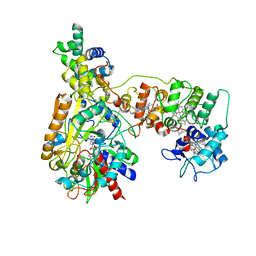 | | Cryo-EM Structure of Membrane-bound Fructose Dehydrogenase from Gluconobacter japonicus variant-N1146Q | | 分子名称: | FE3-S4 CLUSTER, FLAVIN-ADENINE DINUCLEOTIDE, Fructose dehydrogenase cytochrome subunit, ... | | 著者 | Fukawa, E, Miyata, T, Makino, F, Adachi, T, Suzuki, Y, Tanaka, H, Namba, K, Sowa, K, Kitazumi, Y, Shirai, O. | | 登録日 | 2023-12-09 | | 公開日 | 2024-05-22 | | 実験手法 | ELECTRON MICROSCOPY (3.08 Å) | | 主引用文献 | Structural and electrochemical elucidation of biocatalytic mechanisms in direct electron transfer-type D-fructose dehydrogenase.
Electrochim Acta, 490, 2024
|
|
8XCK
 
 | |
8XCJ
 
 | |
8XCI
 
 | |
8XCG
 
 | |
8XC6
 
 | |
8XC4
 
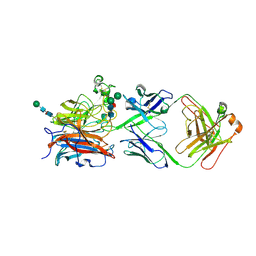 | | Nipah virus attachment glycoprotein head domain in complex with a broadly neutralizing antibody 1E5 | | 分子名称: | 1E5-VH, 1E5-VL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Fan, P.F, Yu, C.M, Chen, W. | | 登録日 | 2023-12-08 | | 公開日 | 2024-01-24 | | 最終更新日 | 2024-08-07 | | 実験手法 | X-RAY DIFFRACTION (3.24 Å) | | 主引用文献 | A potent Henipavirus cross-neutralizing antibody reveals a dynamic fusion-triggering pattern of the G-tetramer.
Nat Commun, 15, 2024
|
|
8XBY
 
 | | The cryo-EM structure of the RAD51 L1 and L2 loops bound to the linker DNA with the blunt end of the nucleosome | | 分子名称: | DNA (5'-D(P*AP*AP*CP*GP*AP*AP*AP*AP*CP*GP*GP*CP*CP*AP*CP*CP*AP*CP*G)-3'), DNA (5'-D(P*CP*GP*TP*GP*GP*TP*GP*GP*CP*CP*GP*TP*TP*TP*TP*CP*GP*TP*T)-3'), DNA repair protein RAD51 homolog 1 | | 著者 | Shioi, T, Hatazawa, S, Ogasawara, M, Takizawa, Y, Kurumizaka, H. | | 登録日 | 2023-12-07 | | 公開日 | 2024-03-27 | | 最終更新日 | 2024-05-08 | | 実験手法 | ELECTRON MICROSCOPY (7.8 Å) | | 主引用文献 | Cryo-EM structures of RAD51 assembled on nucleosomes containing a DSB site.
Nature, 628, 2024
|
|
8XBX
 
 | | The cryo-EM structure of the RAD51 L2 loop bound to the linker DNA with the blunt end of the nucleosome | | 分子名称: | DNA (5'-D(P*AP*AP*CP*GP*AP*AP*AP*AP*CP*GP*GP*CP*CP*AP*CP*CP*AP*CP*G)-3'), DNA (5'-D(P*CP*GP*TP*GP*GP*TP*GP*GP*CP*CP*GP*TP*TP*TP*TP*CP*GP*TP*T)-3'), DNA repair protein RAD51 homolog 1 | | 著者 | Shioi, T, Hatazawa, S, Ogasawara, M, Takizawa, Y, Kurumizaka, H. | | 登録日 | 2023-12-07 | | 公開日 | 2024-03-27 | | 最終更新日 | 2024-04-17 | | 実験手法 | ELECTRON MICROSCOPY (4.36 Å) | | 主引用文献 | Cryo-EM structures of RAD51 assembled on nucleosomes containing a DSB site.
Nature, 628, 2024
|
|
8XBW
 
 | | The cryo-EM structure of the RAD51 N-terminal lobe domain bound to the histone H4 tail of the nucleosome | | 分子名称: | DNA (5'-D(P*AP*CP*CP*GP*CP*TP*TP*AP*AP*AP*CP*GP*CP*AP*CP*GP*TP*A)-3'), DNA (5'-D(P*TP*AP*CP*GP*TP*GP*CP*GP*TP*TP*TP*AP*AP*GP*CP*GP*GP*T)-3'), DNA repair protein RAD51 homolog 1, ... | | 著者 | Shioi, T, Hatazawa, S, Ogasawara, M, Takizawa, Y, Kurumizaka, H. | | 登録日 | 2023-12-07 | | 公開日 | 2024-03-27 | | 最終更新日 | 2024-05-08 | | 実験手法 | ELECTRON MICROSCOPY (2.89 Å) | | 主引用文献 | Cryo-EM structures of RAD51 assembled on nucleosomes containing a DSB site.
Nature, 628, 2024
|
|
8XBV
 
 | | The cryo-EM structure of the RAD51 L1 and L2 loops bound to the linker DNA with the sticky end of the nucleosome | | 分子名称: | DNA (5'-D(P*CP*GP*AP*AP*AP*AP*CP*GP*GP*CP*CP*AP*CP*CP*A)-3'), DNA (5'-D(P*TP*GP*GP*CP*CP*GP*TP*TP*TP*TP*CP*G)-3'), DNA repair protein RAD51 homolog 1 | | 著者 | Shioi, T, Hatazawa, S, Ogasawara, M, Takizawa, Y, Kurumizaka, H. | | 登録日 | 2023-12-07 | | 公開日 | 2024-03-27 | | 最終更新日 | 2024-05-08 | | 実験手法 | ELECTRON MICROSCOPY (7.61 Å) | | 主引用文献 | Cryo-EM structures of RAD51 assembled on nucleosomes containing a DSB site.
Nature, 628, 2024
|
|
8XBU
 
 | | The cryo-EM structure of the decameric RAD51 ring bound to the nucleosome with the linker DNA binding | | 分子名称: | DNA (153-MER), DNA (156-MER), DNA repair protein RAD51 homolog 1, ... | | 著者 | Shioi, T, Hatazawa, S, Ogasawara, M, Takizawa, Y, Kurumizaka, H. | | 登録日 | 2023-12-07 | | 公開日 | 2024-03-27 | | 最終更新日 | 2024-05-08 | | 実験手法 | ELECTRON MICROSCOPY (4.24 Å) | | 主引用文献 | Cryo-EM structures of RAD51 assembled on nucleosomes containing a DSB site.
Nature, 628, 2024
|
|
8XBT
 
 | | The cryo-EM structure of the octameric RAD51 ring bound to the nucleosome with the linker DNA binding | | 分子名称: | DNA (153-MER), DNA (156-MER), DNA repair protein RAD51 homolog 1, ... | | 著者 | Shioi, T, Hatazawa, S, Ogasawara, M, Takizawa, Y, Kurumizaka, H. | | 登録日 | 2023-12-07 | | 公開日 | 2024-03-27 | | 最終更新日 | 2024-05-08 | | 実験手法 | ELECTRON MICROSCOPY (4.12 Å) | | 主引用文献 | Cryo-EM structures of RAD51 assembled on nucleosomes containing a DSB site.
Nature, 628, 2024
|
|
8XBI
 
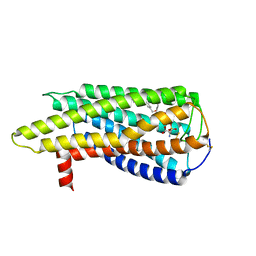 | | Human GPR34 -Gi complex bound to M1, receptor focused | | 分子名称: | (2~{S})-2-azanyl-3-[[(2~{R})-1-ethoxy-3-[3-[2-[(3-phenoxyphenyl)methoxy]phenyl]propanoyloxy]propan-2-yl]oxy-oxidanyl-phosphoryl]oxy-propanoic acid, Probable G-protein coupled receptor 34 | | 著者 | Kawahara, R, Shihoya, W, Nureki, O. | | 登録日 | 2023-12-06 | | 公開日 | 2023-12-27 | | 最終更新日 | 2024-05-15 | | 実験手法 | ELECTRON MICROSCOPY (3.06 Å) | | 主引用文献 | Structural basis for lysophosphatidylserine recognition by GPR34.
Nat Commun, 15, 2024
|
|
8XBH
 
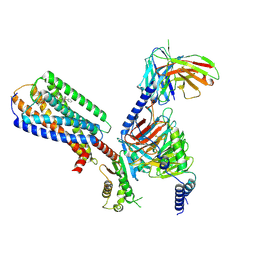 | | Human GPR34 -Gi complex bound to M1 | | 分子名称: | (2~{S})-2-azanyl-3-[[(2~{R})-1-ethoxy-3-[3-[2-[(3-phenoxyphenyl)methoxy]phenyl]propanoyloxy]propan-2-yl]oxy-oxidanyl-phosphoryl]oxy-propanoic acid, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | 著者 | Kawahara, R, Shihoya, W, Nureki, O. | | 登録日 | 2023-12-06 | | 公開日 | 2023-12-27 | | 最終更新日 | 2024-05-15 | | 実験手法 | ELECTRON MICROSCOPY (2.83 Å) | | 主引用文献 | Structural basis for lysophosphatidylserine recognition by GPR34.
Nat Commun, 15, 2024
|
|
8XBG
 
 | |
8XBF
 
 | | Cryo-EM structure of SARS-CoV-2 S-BQ.1 in complex with antibody O5C2 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, O5C2, heavy chain, ... | | 著者 | Hsu, H.F, Wu, M.H, Chang, Y.C, Hsu, S.T.D. | | 登録日 | 2023-12-06 | | 公開日 | 2024-06-19 | | 最終更新日 | 2024-06-26 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | Functional and structural investigation of a broadly neutralizing SARS-CoV-2 antibody.
JCI Insight, 9, 2024
|
|
8XBE
 
 | | Human GPR34 -Gi complex bound to S3E-LysoPS | | 分子名称: | (2~{S})-2-azanyl-3-[[(2~{R})-1-ethoxy-3-[(~{Z})-octadec-9-enoyl]oxy-propan-2-yl]oxy-oxidanyl-phosphoryl]oxy-propanoic acid, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | 著者 | Kawahara, R, Shihoya, W, Nureki, O. | | 登録日 | 2023-12-06 | | 公開日 | 2024-05-15 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Structural basis for lysophosphatidylserine recognition by GPR34.
Nat Commun, 15, 2024
|
|
8XAR
 
 | | Structure-Based Design and Optimization of Methionine Adenosyltransferase 2A (MAT2A) Inhibitors with SAM and Compound 54 | | 分子名称: | 1,2-ETHANEDIOL, 7-chloranyl-2-ethyl-5-pyridin-3-yl-pyrazolo[3,4-c]quinolin-4-one, CHLORIDE ION, ... | | 著者 | Zheng, J.Y, Zhang, G.P, Li, J.J, Tong, S.L. | | 登録日 | 2023-12-05 | | 公開日 | 2024-06-26 | | 実験手法 | X-RAY DIFFRACTION (1.19 Å) | | 主引用文献 | Structure-Based Design and Optimization of Methionine Adenosyltransferase 2A (MAT2A) Inhibitors with High Selectivity, Brain Penetration, and In Vivo Efficacy.
J.Med.Chem., 67, 2024
|
|
