6RD8
 
 | | CryoEM structure of Polytomella F-ATP synthase, c-ring position 2, focussed refinement of Fo and peripheral stalk | | 分子名称: | ASA-10: Polytomella F-ATP synthase associated subunit 10, ASA-9: Polytomella F-ATP synthase associated subunit 9, ATP synthase associated protein ASA1, ... | | 著者 | Murphy, B.J, Klusch, N, Yildiz, O, Kuhlbrandt, W. | | 登録日 | 2019-04-12 | | 公開日 | 2019-07-03 | | 最終更新日 | 2020-09-30 | | 実験手法 | ELECTRON MICROSCOPY (3.08 Å) | | 主引用文献 | Rotary substates of mitochondrial ATP synthase reveal the basis of flexible F 1 -F o coupling.
Science, 364, 2019
|
|
6RDF
 
 | | CryoEM structure of Polytomella F-ATP synthase, Primary rotary state 3, monomer-masked refinement | | 分子名称: | ASA-10: Polytomella F-ATP synthase associated subunit 10, ASA-2: Polytomella F-ATP synthase associated subunit 2, ASA-9: Polytomella F-ATP synthase associated subunit 9, ... | | 著者 | Murphy, B.J, Klusch, N, Yildiz, O, Kuhlbrandt, W. | | 登録日 | 2019-04-12 | | 公開日 | 2019-07-03 | | 最終更新日 | 2024-05-22 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Rotary substates of mitochondrial ATP synthase reveal the basis of flexible F 1 -F o coupling.
Science, 364, 2019
|
|
6RDC
 
 | | CryoEM structure of Polytomella F-ATP synthase, Primary rotary state 2, composite map | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ASA-10: Polytomella F-ATP synthase associated subunit 10, ... | | 著者 | Murphy, B.J, Klusch, N, Yildiz, O, Kuhlbrandt, W. | | 登録日 | 2019-04-12 | | 公開日 | 2019-07-03 | | 最終更新日 | 2024-05-22 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Rotary substates of mitochondrial ATP synthase reveal the basis of flexible F 1 -F o coupling.
Science, 364, 2019
|
|
6RDH
 
 | | CryoEM structure of Polytomella F-ATP synthase, Rotary substate 1A, composite map | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ASA-10: Polytomella F-ATP synthase associated subunit 10, ... | | 著者 | Murphy, B.J, Klusch, N, Yildiz, O, Kuhlbrandt, W. | | 登録日 | 2019-04-12 | | 公開日 | 2019-07-03 | | 最終更新日 | 2024-05-22 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | Rotary substates of mitochondrial ATP synthase reveal the basis of flexible F 1 -F o coupling.
Science, 364, 2019
|
|
6RDO
 
 | | Cryo-EM structure of Polytomella F-ATP synthase, Rotary substate 1C, composite map | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ASA-10: Polytomella F-ATP synthase associated subunit 10, ... | | 著者 | Murphy, B.J, Klusch, N, Yildiz, O, Kuhlbrandt, W. | | 登録日 | 2019-04-12 | | 公開日 | 2019-07-03 | | 最終更新日 | 2024-05-22 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | Rotary substates of mitochondrial ATP synthase reveal the basis of flexible F 1 -F o coupling.
Science, 364, 2019
|
|
6RDY
 
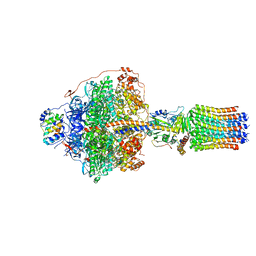 | | Cryo-EM structure of Polytomella F-ATP synthase, Rotary substate 1F, focussed refinement of F1 head and rotor | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase gamma chain, ... | | 著者 | Murphy, B.J, Klusch, N, Yildiz, O, Kuhlbrandt, W. | | 登録日 | 2019-04-12 | | 公開日 | 2019-07-03 | | 最終更新日 | 2024-05-22 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | Rotary substates of mitochondrial ATP synthase reveal the basis of flexible F 1 -F o coupling.
Science, 364, 2019
|
|
6RDW
 
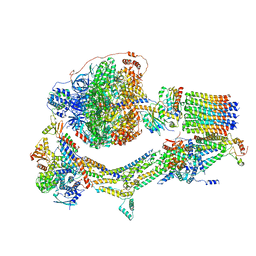 | | Cryo-EM structure of Polytomella F-ATP synthase, Rotary substate 1F, composite map | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ASA-10: Polytomella F-ATP synthase associated subunit 10, ... | | 著者 | Murphy, B.J, Klusch, N, Yildiz, O, Kuhlbrandt, W. | | 登録日 | 2019-04-12 | | 公開日 | 2019-07-03 | | 最終更新日 | 2024-05-22 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | Rotary substates of mitochondrial ATP synthase reveal the basis of flexible F 1 -F o coupling.
Science, 364, 2019
|
|
6RE5
 
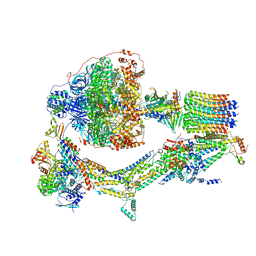 | | Cryo-EM structure of Polytomella F-ATP synthase, Rotary substate 2C, composite map | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ASA-10: Polytomella F-ATP synthase associated subunit 10, ... | | 著者 | Murphy, B.J, Klusch, N, Yildiz, O, Kuhlbrandt, W. | | 登録日 | 2019-04-12 | | 公開日 | 2019-07-03 | | 最終更新日 | 2024-05-22 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Rotary substates of mitochondrial ATP synthase reveal the basis of flexible F 1 -F o coupling.
Science, 364, 2019
|
|
6RED
 
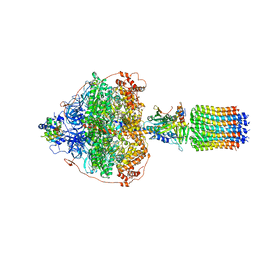 | | Cryo-EM structure of Polytomella F-ATP synthase, Rotary substate 3A, focussed refinement of F1 head and rotor | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase gamma chain, ... | | 著者 | Murphy, B.J, Klusch, N, Yildiz, O, Kuhlbrandt, W. | | 登録日 | 2019-04-12 | | 公開日 | 2019-07-03 | | 最終更新日 | 2024-05-22 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | Rotary substates of mitochondrial ATP synthase reveal the basis of flexible F 1 -F o coupling.
Science, 364, 2019
|
|
6RE9
 
 | | Cryo-EM structure of Polytomella F-ATP synthase, Rotary substate 2D, monomer-masked refinement | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ASA-10: Polytomella F-ATP synthase associated subunit 10, ... | | 著者 | Murphy, B.J, Klusch, N, Yildiz, O, Kuhlbrandt, W. | | 登録日 | 2019-04-12 | | 公開日 | 2019-07-03 | | 最終更新日 | 2024-05-22 | | 実験手法 | ELECTRON MICROSCOPY (3.9 Å) | | 主引用文献 | Rotary substates of mitochondrial ATP synthase reveal the basis of flexible F 1 -F o coupling.
Science, 364, 2019
|
|
6RET
 
 | | Cryo-EM structure of Polytomella F-ATP synthase, Rotary substate 3C, monomer-masked refinement | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ASA-10: Polytomella F-ATP synthase associated subunit 10, ... | | 著者 | Murphy, B.J, Klusch, N, Yildiz, O, Kuhlbrandt, W. | | 登録日 | 2019-04-12 | | 公開日 | 2019-07-03 | | 最終更新日 | 2024-05-22 | | 実験手法 | ELECTRON MICROSCOPY (4.3 Å) | | 主引用文献 | Rotary substates of mitochondrial ATP synthase reveal the basis of flexible F 1 -F o coupling.
Science, 364, 2019
|
|
4E7V
 
 | | The structure of R6 bovine insulin | | 分子名称: | CHLORIDE ION, Insulin A chain, Insulin B chain, ... | | 著者 | Harris, P, Frankaer, C.G, Knudsen, M.V. | | 登録日 | 2012-03-19 | | 公開日 | 2012-04-04 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | The structures of T(6), T(3)R(3) and R(6) bovine insulin: combining X-ray diffraction and absorption spectroscopy.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
6RZU
 
 | | Structure of s-Mgm1 decorating the outer surface of tubulated lipid membranes in the GTPgammaS bound state | | 分子名称: | Putative mitochondrial dynamin protein | | 著者 | Faelber, K, Dietrich, L, Noel, J.K, Sanchez, R, Kudryashev, M, Kuelbrandt, W, Daumke, O. | | 登録日 | 2019-06-13 | | 公開日 | 2019-07-24 | | 最終更新日 | 2020-11-18 | | 実験手法 | ELECTRON MICROSCOPY (14.7 Å) | | 主引用文献 | Structure and assembly of the mitochondrial membrane remodelling GTPase Mgm1.
Nature, 571, 2019
|
|
6RO1
 
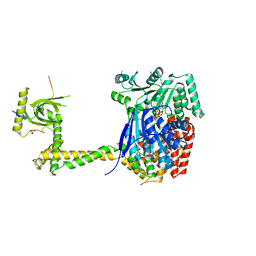 | | X-ray crystal structure of the MTR4 NVL complex | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, Exosome RNA helicase MTR4, ... | | 著者 | Lingaraju, M, Langer, L.M, Basquin, J, Falk, S, Conti, E. | | 登録日 | 2019-05-10 | | 公開日 | 2019-07-03 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (3.07 Å) | | 主引用文献 | The MTR4 helicase recruits nuclear adaptors of the human RNA exosome using distinct arch-interacting motifs.
Nat Commun, 10, 2019
|
|
6S6Y
 
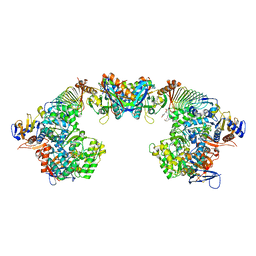 | | X-ray crystal structure of the formyltransferase/hydrolase complex (FhcABCD) from Methylorubrum extorquens in complex with methylofuran | | 分子名称: | (2~{S})-3-[4-[[5-(aminomethyl)furan-3-yl]methoxy]phenyl]-2-(methylamino)propanoic acid, 1,2-ETHANEDIOL, AMINO GROUP, ... | | 著者 | Wagner, T, Hemmann, J.L, Shima, S, Vorholt, J. | | 登録日 | 2019-07-04 | | 公開日 | 2019-12-04 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (3.1 Å) | | 主引用文献 | Methylofuran is a prosthetic group of the formyltransferase/hydrolase complex and shuttles one-carbon units between two active sites.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
4BW8
 
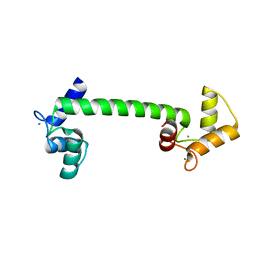 | | Calmodulin with small bend in central helix | | 分子名称: | CALCIUM ION, CALMODULIN | | 著者 | Kursula, P. | | 登録日 | 2013-06-30 | | 公開日 | 2014-01-15 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Crystallographic Snapshots of Initial Steps in the Collapse of the Calmodulin Central Helix
Acta Crystallogr.,Sect.D, 70, 2014
|
|
6RZV
 
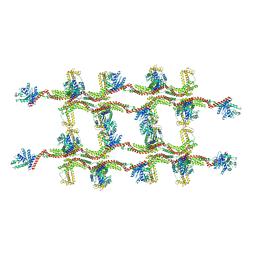 | | Structure of s-Mgm1 decorating the inner surface of tubulated lipid membranes | | 分子名称: | Putative mitochondrial dynamin protein | | 著者 | Faelber, K, Dietrich, L, Noel, J.K, Sanchez, R, Kudryashev, M, Kuelbrandt, W, Daumke, O. | | 登録日 | 2019-06-13 | | 公開日 | 2019-07-24 | | 最終更新日 | 2020-11-18 | | 実験手法 | ELECTRON MICROSCOPY (20.6 Å) | | 主引用文献 | Structure and assembly of the mitochondrial membrane remodelling GTPase Mgm1.
Nature, 571, 2019
|
|
3FAS
 
 | | X-ray structure of iGluR4 flip ligand-binding core (S1S2) in complex with (S)-glutamate at 1.40A resolution | | 分子名称: | GLUTAMIC ACID, GLYCEROL, Glutamate receptor 4, ... | | 著者 | Kasper, C, Frydenvang, K, Naur, P, Gajhede, M, Kastrup, J.S. | | 登録日 | 2008-11-18 | | 公開日 | 2008-12-09 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Molecular mechanism of agonist recognition by the ligand-binding core of the ionotropic glutamate receptor 4
Febs Lett., 582, 2008
|
|
5XYX
 
 | | The structure of p38 alpha in complex with a triazol inhibitor | | 分子名称: | Mitogen-activated protein kinase 14, N-(2-chloro-6-fluorobenzyl)-5-(furan-2-yl)-2H-1,2,4-triazol-3-amine | | 著者 | Wang, Y.L, Sun, Y.Z, Cao, R, Liu, D, Li, L, Qi, X.B, Huang, N. | | 登録日 | 2017-07-11 | | 公開日 | 2018-01-17 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.61 Å) | | 主引用文献 | In Silico Identification of a Novel Hinge-Binding Scaffold for Kinase Inhibitor Discovery.
J. Med. Chem., 60, 2017
|
|
6RZW
 
 | | Structure of s-Mgm1 decorating the inner surface of tubulated lipid membranes in the GTPgammaS bound state | | 分子名称: | Putative mitochondrial dynamin protein | | 著者 | Faelber, K, Dietrich, L, Noel, J.K, Sanchez, R, Kudryashev, M, Kuelbrandt, W, Daumke, O. | | 登録日 | 2019-06-13 | | 公開日 | 2019-07-24 | | 最終更新日 | 2020-11-18 | | 実験手法 | ELECTRON MICROSCOPY (18.799999 Å) | | 主引用文献 | Structure and assembly of the mitochondrial membrane remodelling GTPase Mgm1.
Nature, 571, 2019
|
|
5WJJ
 
 | | Structure-based Design, Synthesis, and Biological Evaluation of Imidazo[1,2-b]pyridazine-based p38 MAP Kinase Inhibitors | | 分子名称: | Mitogen-activated protein kinase 14, N-{4-[2-(4-fluoro-3-methylphenyl)imidazo[1,2-b]pyridazin-3-yl]pyridin-2-yl}-2-methyl-1-oxo-1lambda~5~-pyridine-4-carboxamide | | 著者 | Snell, G.P, Okada, K, Bragstad, K, Sang, B.-C. | | 登録日 | 2017-07-23 | | 公開日 | 2018-01-17 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Structure-based design, synthesis, and biological evaluation of imidazo[1,2-b]pyridazine-based p38 MAP kinase inhibitors.
Bioorg. Med. Chem., 26, 2018
|
|
5XYY
 
 | | The structure of p38 alpha in complex with a triazol inhibitor | | 分子名称: | 3-(5-{[(2-chloro-6-fluorophenyl)methyl]amino}-4H-1,2,4-triazol-3-yl)phenol, Mitogen-activated protein kinase 14 | | 著者 | Wang, Y.L, Sun, Y.Z, Cao, R, Liu, D, Li, L, Qi, X.B, Huang, N. | | 登録日 | 2017-07-11 | | 公開日 | 2018-01-17 | | 最終更新日 | 2024-04-10 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | In Silico Identification of a Novel Hinge-Binding Scaffold for Kinase Inhibitor Discovery.
J. Med. Chem., 60, 2017
|
|
4E7T
 
 | | The structure of T6 bovine insulin | | 分子名称: | Insulin A chain, Insulin B chain, ZINC ION | | 著者 | Harris, P, Frankaer, C.G, Knudsen, M.V. | | 登録日 | 2012-03-19 | | 公開日 | 2012-04-04 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | The structures of T(6), T(3)R(3) and R(6) bovine insulin: combining X-ray diffraction and absorption spectroscopy.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
6SHD
 
 | |
3G10
 
 | | Structure of S. pombe Pop2p - Mg2+ and Mn2+ bound form | | 分子名称: | CCR4-Not complex subunit Caf1, MAGNESIUM ION, MANGANESE (II) ION | | 著者 | Andersen, K.R, Jonstrup, A.T, Van, L.B, Brodersen, D.E. | | 登録日 | 2009-01-29 | | 公開日 | 2009-03-31 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.597 Å) | | 主引用文献 | The activity and selectivity of fission yeast Pop2p are affected by a high affinity for Zn2+ and Mn2+ in the active site
Rna, 15, 2009
|
|
