2F5O
 
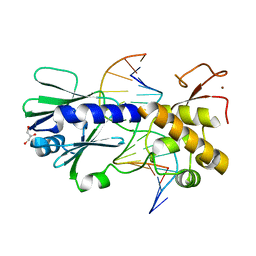 | | MutM crosslinked to undamaged DNA sampling G:C base pair IC3 | | 分子名称: | 5'-D(*AP*GP*GP*TP*AP*GP*AP*TP*CP*CP*GP*GP*AP*CP*GP*C)-3', 5'-D(*TP*GP*C*GP*TP*CP*CP*GP*GP*AP*TP*CP*TP*AP*CP*C)-3', GLYCEROL, ... | | 著者 | Banerjee, A, Santos, W.L, Verdine, G.L. | | 登録日 | 2005-11-26 | | 公開日 | 2006-03-07 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (2.05 Å) | | 主引用文献 | Structure of a DNA glycosylase searching for lesions.
Science, 311, 2006
|
|
2F6D
 
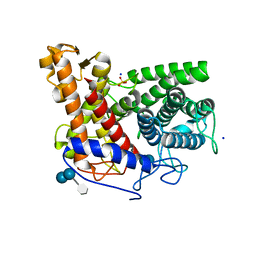 | | Structure of the complex of a glucoamylase from Saccharomycopsis fibuligera with acarbose | | 分子名称: | 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, Glucoamylase GLU1, PHOSPHATE ION, ... | | 著者 | Sevcik, J, Hostinova, E, Solovicova, A, Gasperik, J, Dauter, Z, Wilson, K.S. | | 登録日 | 2005-11-29 | | 公開日 | 2006-05-23 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Structure of the complex of a yeast glucoamylase with acarbose reveals the presence of a raw starch binding site on the catalytic domain.
Febs J., 273, 2006
|
|
2F7N
 
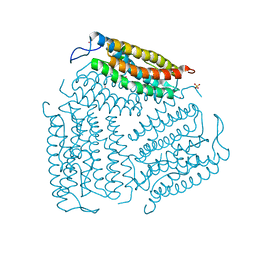 | | Structure of D. radiodurans Dps-1 | | 分子名称: | COBALT (II) ION, DNA-binding stress response protein, Dps family, ... | | 著者 | Lee, Y.H, Kim, S.G, Bhattacharyya, G, Grove, A. | | 登録日 | 2005-12-01 | | 公開日 | 2006-11-14 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Crystal structure of Dps-1, a functionally distinct Dps protein from Deinococcus radiodurans.
J.Mol.Biol., 361, 2006
|
|
255L
 
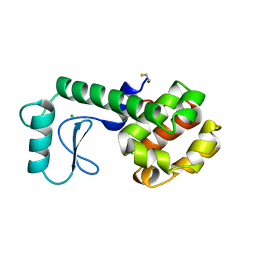 | | HYDROLASE | | 分子名称: | BETA-MERCAPTOETHANOL, CHLORIDE ION, LYSOZYME | | 著者 | Kuroki, R, Shoichet, B, Weaver, L.H, Matthews, B.W. | | 登録日 | 1997-11-10 | | 公開日 | 1998-01-28 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | A relationship between protein stability and protein function.
Proc.Natl.Acad.Sci.USA, 92, 1995
|
|
210L
 
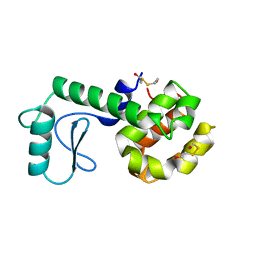 | | PROTEIN STRUCTURE PLASTICITY EXEMPLIFIED BY INSERTION AND DELETION MUTANTS IN T4 LYSOZYME | | 分子名称: | 2-HYDROXYETHYL DISULFIDE, T4 LYSOZYME | | 著者 | Vetter, I.R, Baase, W.A, Heinz, D.W, Xiong, J.-P, Snow, S, Matthews, B.W. | | 登録日 | 1996-09-23 | | 公開日 | 1996-12-23 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.89 Å) | | 主引用文献 | Protein structural plasticity exemplified by insertion and deletion mutants in T4 lysozyme.
Protein Sci., 5, 1996
|
|
2FHC
 
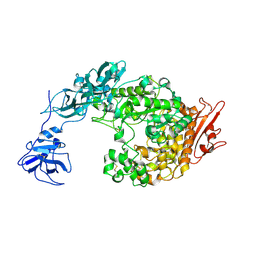 | | Crystal Structure Analysis of Klebsiella pneumoniae pullulanase complexed with maltotriose | | 分子名称: | Alpha-dextrin endo-1,6-alpha-glucosidase, CALCIUM ION, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, ... | | 著者 | Mikami, B, Iwamoto, H, Katsuya, Y, Yoon, H.-J, Demirkan-Sarikaya, E, Malle, D. | | 登録日 | 2005-12-23 | | 公開日 | 2006-06-13 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Crystal structure of pullulanase: evidence for parallel binding of oligosaccharides in the active site
J.Mol.Biol., 359, 2006
|
|
2AC1
 
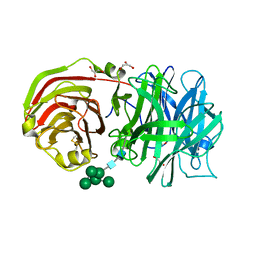 | | Crystal structure of a cell-wall invertase from Arabidopsis thaliana | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, ... | | 著者 | Verhaest, M, Le Roy, K, De Ranter, C, Van Laere, A, Van den Ende, W, Rabijns, A. | | 登録日 | 2005-07-18 | | 公開日 | 2006-08-29 | | 最終更新日 | 2024-10-30 | | 実験手法 | X-RAY DIFFRACTION (2.15 Å) | | 主引用文献 | X-ray diffraction structure of a cell-wall invertase from Arabidopsis thaliana.
Acta Crystallogr.,Sect.D, 62, 2006
|
|
223L
 
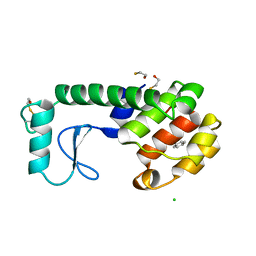 | | GENERATING LIGAND BINDING SITES IN T4 LYSOZYME USING DEFICIENCY-CREATING SUBSTITUTIONS | | 分子名称: | BENZENE, BETA-MERCAPTOETHANOL, CHLORIDE ION, ... | | 著者 | Baldwin, E.P, Baase, W.A, Zhang, X.-J, Feher, V, Matthews, B.W. | | 登録日 | 1997-06-25 | | 公開日 | 1998-03-18 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Generation of ligand binding sites in T4 lysozyme by deficiency-creating substitutions.
J.Mol.Biol., 277, 1998
|
|
258L
 
 | | AN ADAPTABLE METAL-BINDING SITE ENGINEERED INTO T4 LYSOZYME | | 分子名称: | CHLORIDE ION, LYSOZYME, ZINC ION | | 著者 | Wray, J.W, Baase, W.A, Ostheimer, G.J, Matthews, B.W. | | 登録日 | 1999-01-05 | | 公開日 | 2000-09-11 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Use of a non-rigid region in T4 lysozyme to design an adaptable metal-binding site.
Protein Eng., 13, 2000
|
|
2FGL
 
 | | An alkali thermostable F/10 xylanase from alkalophilic Bacillus sp. NG-27 | | 分子名称: | MAGNESIUM ION, alkaline thermostable endoxylanase, alpha-D-xylopyranose, ... | | 著者 | Ramakumar, S, Manikandan, K, Bhardwaj, A, Reddy, V.S, Lokanath, N.K, Ghosh, A. | | 登録日 | 2005-12-22 | | 公開日 | 2006-09-26 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Crystal structures of native and xylosaccharide-bound alkali thermostable xylanase from an alkalophilic Bacillus sp. NG-27: structural insights into alkalophilicity and implications for adaptation to polyextreme conditions.
Protein Sci., 15, 2006
|
|
2AEY
 
 | | Crystal structure of fructan 1-exohydrolase IIa from Cichorium intybus in complex with 2,5 dideoxy-2,5-immino-D-mannitol | | 分子名称: | 2,5-DIDEOXY-2,5-IMINO-D-MANNITOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, alpha-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Verhaest, M, Le Roy, K, De Ranter, C.J, Van Laere, A, Van den Ende, W, Rabijns, A. | | 登録日 | 2005-07-25 | | 公開日 | 2006-08-29 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (3.27 Å) | | 主引用文献 | Insights into the fine architecture of the active site of chicory fructan 1-exohydrolase: 1-kestose as substrate vs sucrose as inhibitor.
New Phytol, 174, 2007
|
|
2GZ2
 
 | | Structure of Aspartate Semialdehyde Dehydrogenase (ASADH) from Streptococcus pneumoniae complexed with 2',5'-ADP | | 分子名称: | ADENOSINE-2'-5'-DIPHOSPHATE, Aspartate beta-semialdehyde dehydrogenase | | 著者 | Faehnle, C.R, Le Coq, J, Liu, X, Viola, R.E. | | 登録日 | 2006-05-10 | | 公開日 | 2006-08-15 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Examination of key intermediates in the catalytic cycle of aspartate-beta-semialdehyde dehydrogenase from a gram-positive infectious bacteria.
J.Biol.Chem., 281, 2006
|
|
2HBU
 
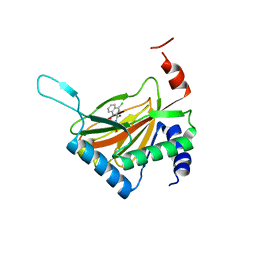 | | Crystal structure of HIF prolyl hydroxylase EGLN-1 in complex with a biologically active inhibitor | | 分子名称: | Egl nine homolog 1, FE (II) ION, N-[(1-CHLORO-4-HYDROXYISOQUINOLIN-3-YL)CARBONYL]GLYCINE | | 著者 | Evdokimov, A.G, Walter, R.L, Mekel, M, Pokross, M.E, Kawamoto, R, Boyer, A. | | 登録日 | 2006-06-14 | | 公開日 | 2006-06-27 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Crystal structure of HIF prolyl hydroxylase in complex with a biologically active inhibitor
To be Published
|
|
1ZWN
 
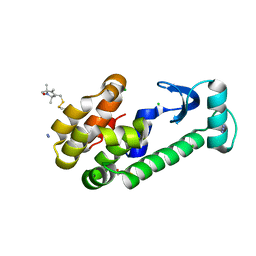 | | Crystal structure of spin labeled T4 Lysozyme (V131R1B) | | 分子名称: | 2-HYDROXYETHYL DISULFIDE, AZIDE ION, CHLORIDE ION, ... | | 著者 | Fleissner, M.R, Cascio, D, Sawaya, M.R, Hideg, K, Hubbell, W.L. | | 登録日 | 2005-06-03 | | 公開日 | 2006-10-17 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Crystal structure of spin labeled T4 Lysozyme (V131R1B)
To be Published
|
|
2EW8
 
 | |
2EXJ
 
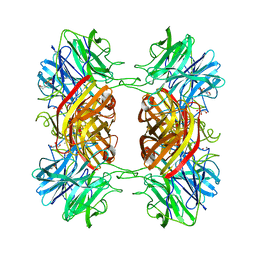 | | Structure of the family43 beta-Xylosidase D128G mutant from geobacillus stearothermophilus in complex with xylobiose | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, GLYCEROL, ... | | 著者 | Brux, C, Niefind, K, Shallom-Shezifi, D, Shoham, Y, Schomburg, D. | | 登録日 | 2005-11-08 | | 公開日 | 2006-04-04 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | The Structure of an Inverting GH43 beta-Xylosidase from Geobacillus stearothermophilus with its Substrate Reveals the Role of the Three Catalytic Residues.
J.Mol.Biol., 359, 2006
|
|
2FLI
 
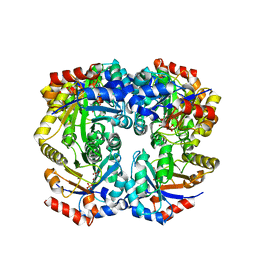 | | The crystal structure of D-ribulose 5-phosphate 3-epimerase from Streptococus pyogenes complexed with D-xylitol 5-phosphate | | 分子名称: | D-XYLITOL-5-PHOSPHATE, ZINC ION, ribulose-phosphate 3-epimerase | | 著者 | Fedorov, A.A, Fedorov, E.V, Akana, J, Gerlt, J.A, Almo, S.C. | | 登録日 | 2006-01-06 | | 公開日 | 2006-03-07 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | d-Ribulose 5-Phosphate 3-Epimerase: Functional and Structural Relationships to Members of the Ribulose-Phosphate Binding (beta/alpha)(8)-Barrel Superfamily(,).
Biochemistry, 45, 2006
|
|
2JG7
 
 | | Crystal structure of Seabream Antiquitin and Elucidation of its substrate specificity | | 分子名称: | ANTIQUITIN, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | 著者 | Tang, W.K, Wong, K.B, Cha, S.S, Lee, H.S, Cheng, C.H.K, Fong, W.P. | | 登録日 | 2007-02-09 | | 公開日 | 2008-05-13 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2.83 Å) | | 主引用文献 | The Crystal Structure of Seabream Antiquitin Reveals the Structural Basis of its Substrate Specificity.
FEBS Lett., 582, 2008
|
|
2F1A
 
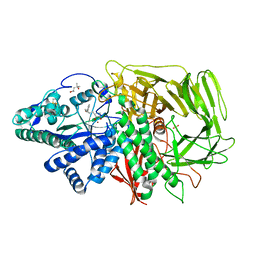 | | GOLGI ALPHA-MANNOSIDASE II COMPLEX WITH (2R,3R,4S)-2-({[(1S)-2-hydroxy-1-phenylethyl]amino}methyl)pyrrolidine-3,4-diol | | 分子名称: | (2R,3R,4S)-2-({[(1S)-2-HYDROXY-1-PHENYLETHYL]AMINO}METHYL)PYRROLIDINE-3,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Kuntz, D.A, Rose, D.R. | | 登録日 | 2005-11-14 | | 公開日 | 2006-12-05 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (1.45 Å) | | 主引用文献 | Evaluation of docking programs for predicting binding of Golgi alpha-mannosidase II inhibitors: a comparison with crystallography.
Proteins, 69, 2007
|
|
2A1O
 
 | |
2A7F
 
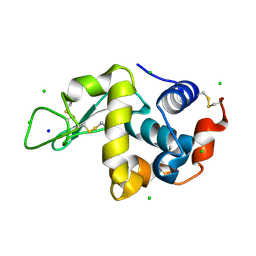 | | On the Routine Use of Soft X-Rays in Macromolecular Crystallography, Part III- The Optimal Data Collection Wavelength | | 分子名称: | CHLORIDE ION, Lysozyme C, SODIUM ION | | 著者 | Mueller-Dieckmann, C, Panjikar, S, Tucker, P.A, Weiss, M.S. | | 登録日 | 2005-07-05 | | 公開日 | 2005-07-19 | | 最終更新日 | 2024-10-30 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | On the routine use of soft X-rays in macromolecular crystallography. Part III. The optimal data-collection wavelength.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
2A8V
 
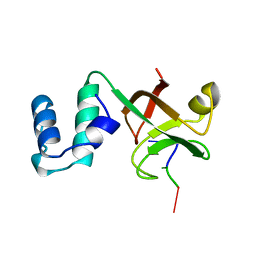 | | RHO TRANSCRIPTION TERMINATION FACTOR/RNA COMPLEX | | 分子名称: | 5'-R(P*CP*CP*C)-3', 5'-R(P*CP*CP*CP*CP*CP*C)-3', RNA BINDING DOMAIN OF RHO TRANSCRIPTION TERMINATION FACTOR | | 著者 | Bogden, C.E, Fass, D, Bergman, N, Nichols, M.D, Berger, J.M. | | 登録日 | 1998-11-08 | | 公開日 | 1999-04-26 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | The structural basis for terminator recognition by the Rho transcription termination factor.
Mol.Cell, 3, 1999
|
|
2EXK
 
 | | Structure of the family43 beta-Xylosidase E187G from geobacillus stearothermophilus in complex with xylobiose | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, GLYCEROL, ... | | 著者 | Brux, C, Niefind, K, Shallom-Shezifi, D, Shoham, Y, Schomburg, D. | | 登録日 | 2005-11-08 | | 公開日 | 2006-04-04 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | The Structure of an Inverting GH43 beta-Xylosidase from Geobacillus stearothermophilus with its Substrate Reveals the Role of the Three Catalytic Residues.
J.Mol.Biol., 359, 2006
|
|
2A7D
 
 | | On the Routine Use of Soft X-Rays in Macromolecular Crystallography, Part III- The Optimal Data Collection Wavelength | | 分子名称: | CHLORIDE ION, Lysozyme C, SODIUM ION, ... | | 著者 | Mueller-Dieckmann, C, Panjikar, S, Tucker, P.A, Weiss, M.S. | | 登録日 | 2005-07-05 | | 公開日 | 2005-07-19 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (1.66 Å) | | 主引用文献 | On the routine use of soft X-rays in macromolecular crystallography. Part III. The optimal data-collection wavelength.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
2FCO
 
 | | Crystal Structure of Bacillus stearothermophilus PrfA-Holliday Junction Resolvase | | 分子名称: | 1,2-ETHANEDIOL, MAGNESIUM ION, recombination protein U (penicillin-binding protein related factor A) | | 著者 | Li, J, Jedrzejas, M.J. | | 登録日 | 2005-12-12 | | 公開日 | 2006-11-21 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Structure, flexibility, and mechanism of the Bacillus stearothermophilus RecU Holliday junction resolvase.
Proteins, 68, 2007
|
|
