4JW1
 
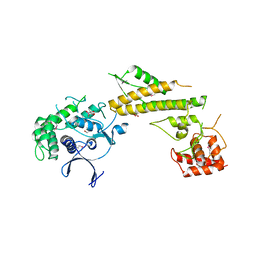 | | Crystal structure of N-terminal 618-residue fragment of LepB from Legionella pneumophila | | 分子名称: | CITRATE ANION, Effector protein B, GLYCEROL | | 著者 | Hu, L, Yao, Q, Zhu, Y, Shao, F. | | 登録日 | 2013-03-26 | | 公開日 | 2013-05-08 | | 最終更新日 | 2013-08-14 | | 実験手法 | X-RAY DIFFRACTION (3.16 Å) | | 主引用文献 | Structural analyses of Legionella LepB reveal a new GAP fold that catalytically mimics eukaryotic RasGAP
Cell Res., 23, 2013
|
|
1RSC
 
 | | STRUCTURE OF AN EFFECTOR INDUCED INACTIVATED STATE OF RIBULOSE BISPHOSPHATE CARBOXYLASE(SLASH)OXYGENASE: THE BINARY COMPLEX BETWEEN ENZYME AND XYLULOSE BISPHOSPHATE | | 分子名称: | RIBULOSE 1,5 BISPHOSPHATE CARBOXYLASE/OXYGENASE (LARGE CHAIN), RIBULOSE 1,5 BISPHOSPHATE CARBOXYLASE/OXYGENASE (SMALL CHAIN), XYLULOSE-1,5-BISPHOSPHATE | | 著者 | Newman, J, Gutteridge, S. | | 登録日 | 1994-03-29 | | 公開日 | 1995-05-08 | | 最終更新日 | 2024-06-05 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Structure of an effector-induced inactivated state of ribulose 1,5-bisphosphate carboxylase/oxygenase: the binary complex between enzyme and xylulose 1,5-bisphosphate.
Structure, 2, 1994
|
|
6RJR
 
 | | Crystal structure of a Fungal Catalase at 1.9 Angstrom | | 分子名称: | CHLORIDE ION, Catalase, GLYCEROL, ... | | 著者 | Gomez, S, Navas-Yuste, S, Payne, A.M, Rivera, W, Lopez-Estepa, M, Brangbour, C, Fulla, D, Juanhuix, J, Fernandez, F.J, Vega, M.C. | | 登録日 | 2019-04-29 | | 公開日 | 2020-03-11 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.895 Å) | | 主引用文献 | Peroxisomal catalases from the yeasts Pichia pastoris and Kluyveromyces lactis as models for oxidative damage in higher eukaryotes.
Free Radic. Biol. Med., 141, 2019
|
|
4JVS
 
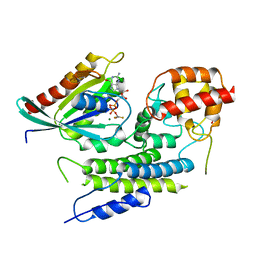 | | Crystal structure of LepB GAP domain from Legionella drancourtii in complex with Rab1-GDP and AlF3 | | 分子名称: | ACETIC ACID, ALUMINUM FLUORIDE, GUANOSINE-5'-DIPHOSPHATE, ... | | 著者 | Yu, Q, Yao, Q, Wang, D.-C, Shao, F. | | 登録日 | 2013-03-26 | | 公開日 | 2013-05-08 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.783 Å) | | 主引用文献 | Structural analyses of Legionella LepB reveal a new GAP fold that catalytically mimics eukaryotic RasGAP
Cell Res., 23, 2013
|
|
7R5M
 
 | |
3E20
 
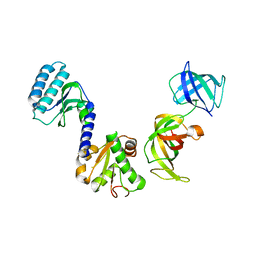 | | Crystal structure of S.pombe eRF1/eRF3 complex | | 分子名称: | Eukaryotic peptide chain release factor GTP-binding subunit, Eukaryotic peptide chain release factor subunit 1 | | 著者 | Cheng, Z, Lim, M, Kong, C, Song, H. | | 登録日 | 2008-08-05 | | 公開日 | 2009-05-19 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (3.5 Å) | | 主引用文献 | Structural insights into eRF3 and stop codon recognition by eRF1
Genes Dev., 23, 2009
|
|
4U1C
 
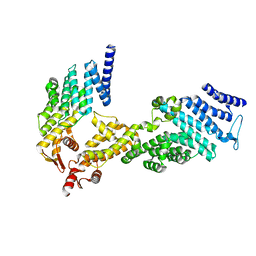 | |
6RJN
 
 | | Crystal structure of a Fungal Catalase at 2.3 Angstroms | | 分子名称: | CHLORIDE ION, Catalase, DI(HYDROXYETHYL)ETHER, ... | | 著者 | Gomez, S, Navas-Yuste, S, Payne, A.M, Rivera, W, Lopez-Estepa, M, Brangbour, C, Fulla, D, Juanhuix, J, Fernandez, F.J, Vega, M.C. | | 登録日 | 2019-04-28 | | 公開日 | 2020-03-04 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.295 Å) | | 主引用文献 | Peroxisomal catalases from the yeasts Pichia pastoris and Kluyveromyces lactis as models for oxidative damage in higher eukaryotes.
Free Radic. Biol. Med., 141, 2019
|
|
6EZP
 
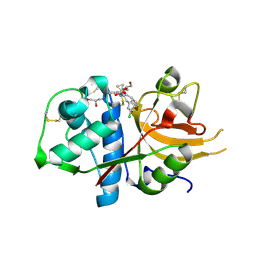 | | CATHEPSIN L IN COMPLEX WITH (3S,14E)-19-chloro-N-(1-cyanocyclopropyl)-5-oxo-12,17-dioxa-4-azatricyclo[16.2.2.06,11]docosa-1(21),6(11),7,9,14,18(22),19-heptaene-3-carboxamide | | 分子名称: | (3~{S},14~{E})-19-chloranyl-~{N}-(1-cyanocyclopropyl)-5-oxidanylidene-12,17-dioxa-4-azatricyclo[16.2.2.0^{6,11}]docosa-1(21),6(11),7,9,14,18(22),19-heptaene-3-carboxamide, Cathepsin L1, GLYCEROL | | 著者 | Banner, D.W, Benz, J, Kuglstatter, A. | | 登録日 | 2017-11-16 | | 公開日 | 2018-04-11 | | 最終更新日 | 2018-05-09 | | 実験手法 | X-RAY DIFFRACTION (1.37 Å) | | 主引用文献 | Repurposing a Library of Human Cathepsin L Ligands: Identification of Macrocyclic Lactams as Potent Rhodesain and Trypanosoma brucei Inhibitors.
J. Med. Chem., 61, 2018
|
|
6EXO
 
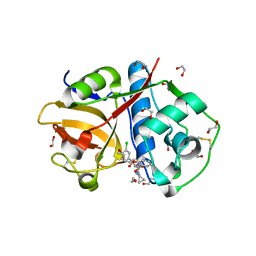 | | Crystal Structure of Rhodesain in complex with a Macrolactam Inhibitor | | 分子名称: | (3~{S},14~{E})-19-chloranyl-~{N}-(1-cyanocyclopropyl)-5-oxidanylidene-12,17-dioxa-4-azatricyclo[16.2.2.0^{6,11}]docosa-1(21),6(11),7,9,14,18(22),19-heptaene-3-carboxamide, 1,2-ETHANEDIOL, Cysteine protease | | 著者 | Dietzel, U, Kisker, C. | | 登録日 | 2017-11-08 | | 公開日 | 2018-04-11 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Repurposing a Library of Human Cathepsin L Ligands: Identification of Macrocyclic Lactams as Potent Rhodesain and Trypanosoma brucei Inhibitors.
J. Med. Chem., 61, 2018
|
|
6EZX
 
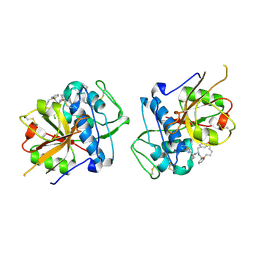 | | CATHEPSIN L IN COMPLEX WITH (3S,14E)-19-chloro-N-(1-cyanocyclopropyl)-5-oxo-17-oxa-4-azatricyclo[16.2.2.06,11]docosa-1(21),6,8,10,14,18(22),19-heptaene-3-carboxamide | | 分子名称: | (3~{S},14~{E})-19-chloranyl-~{N}-[1-(iminomethyl)cyclopropyl]-5-oxidanylidene-17-oxa-4-azatricyclo[16.2.2.0^{6,11}]docosa-1(21),6,8,10,14,18(22),19-heptaene-3-carboxamide, Cathepsin L1 | | 著者 | Banner, D.W, Benz, J, Kuglstatter, A. | | 登録日 | 2017-11-16 | | 公開日 | 2018-04-11 | | 最終更新日 | 2018-05-09 | | 実験手法 | X-RAY DIFFRACTION (2.34 Å) | | 主引用文献 | Repurposing a Library of Human Cathepsin L Ligands: Identification of Macrocyclic Lactams as Potent Rhodesain and Trypanosoma brucei Inhibitors.
J. Med. Chem., 61, 2018
|
|
6EX8
 
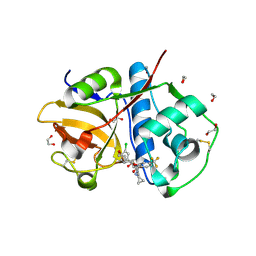 | | Crystal Structure of Rhodesain in complex with a Macrolactam Inhibitor | | 分子名称: | (3~{S})-~{N}-[1-(aminomethyl)cyclopropyl]-19-chloranyl-5-oxidanylidene-9-(trifluoromethyl)-12,17-dioxa-4-azatricyclo[16.2.2.0^{6,11}]docosa-1(20),6(11),7,9,18,21-hexaene-3-carboxamide, 1,2-ETHANEDIOL, Cysteine protease | | 著者 | Dietzel, U, Kisker, C. | | 登録日 | 2017-11-07 | | 公開日 | 2018-04-11 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Repurposing a Library of Human Cathepsin L Ligands: Identification of Macrocyclic Lactams as Potent Rhodesain and Trypanosoma brucei Inhibitors.
J. Med. Chem., 61, 2018
|
|
1LP4
 
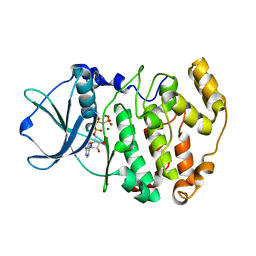 | | Crystal structure of a binary complex of the catalytic subunit of protein kinase CK2 with Mg-AMPPNP | | 分子名称: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, Protein kinase CK2 | | 著者 | Niefind, K, Puetter, M, Guerra, B, Issinger, O.-G, Schomburg, D. | | 登録日 | 2002-05-07 | | 公開日 | 2002-05-29 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.86 Å) | | 主引用文献 | Inclining the purine base binding plane in protein kinase CK2 by exchanging the flanking side-chains generates a preference for ATP as a cosubstrate.
J.Mol.Biol., 347, 2005
|
|
8OTM
 
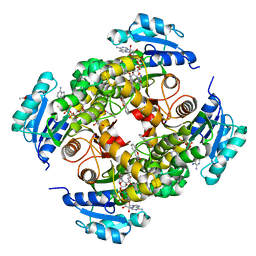 | | structure of InhA from mycobacterium tuberculosis in complex with N-((1-(3-hydroxy-4-phenoxybenzyl)-1H-1,2,3-triazol-4-yl)methyl)-2-oxo-2H-chromene-3-carboxamide | | 分子名称: | 1,2-ETHANEDIOL, 2-oxidanylidene-~{N}-[[1-[(3-oxidanyl-4-phenoxy-phenyl)methyl]-1,2,3-triazol-4-yl]methyl]chromene-3-carboxamide, ACETATE ION, ... | | 著者 | Chebaiki, M, Maveyraud, L, Tamhaev, R, Lherbet, C, Mourey, L. | | 登録日 | 2023-04-21 | | 公開日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Discovery of new diaryl ether inhibitors against Mycobacterium tuberculosis targeting the minor portal of InhA.
Eur.J.Med.Chem., 259, 2023
|
|
1QVO
 
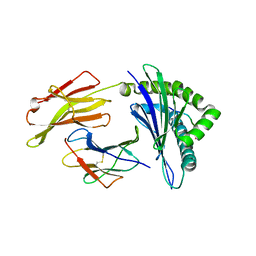 | | STRUCTURES OF HLA-A*1101 IN COMPLEX WITH IMMUNODOMINANT NONAMER AND DECAMER HIV-1 EPITOPES CLEARLY REVEAL THE PRESENCE OF A MIDDLE ANCHOR RESIDUE | | 分子名称: | Beta-2-microglobulin, HLA class I histocompatibility antigen, A-11 alpha chain, ... | | 著者 | Li, L, McNicholl, J.M, Bouvier, M. | | 登録日 | 2003-08-28 | | 公開日 | 2004-06-01 | | 最終更新日 | 2020-02-19 | | 実験手法 | X-RAY DIFFRACTION (2.22 Å) | | 主引用文献 | Structures of HLA-A*1101 complexed with immunodominant nonamer and decamer HIV-1 epitopes clearly reveal the presence of a middle, secondary anchor residue.
J.Immunol., 172, 2004
|
|
8OTN
 
 | | structure of InhA from mycobacterium tuberculosis in complex with inhibitor 7-((1-(3-Hydroxy-4-phenoxybenzyl)-1H-1,2,3-triazol-4-yl)methoxy)-4-methyl-2H-chromen-2-one | | 分子名称: | 4-methyl-7-[[1-[(3-oxidanyl-4-phenoxy-phenyl)methyl]-1,2,3-triazol-4-yl]methoxy]chromen-2-one, Enoyl-[acyl-carrier-protein] reductase [NADH], NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | 著者 | Chebaiki, M, Maveyraud, L, Tamhaev, R, Lherbet, C, Mourey, L. | | 登録日 | 2023-04-21 | | 公開日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (1.962 Å) | | 主引用文献 | Discovery of new diaryl ether inhibitors against Mycobacterium tuberculosis targeting the minor portal of InhA.
Eur.J.Med.Chem., 259, 2023
|
|
8OTL
 
 | | structure of InhA from Mycobacterium tuberculosis in complex with 5-(((4-(2-hydroxyphenoxy)benzyl)(octyl)amino)methyl)-2-phenoxyphenol | | 分子名称: | 1,2-ETHANEDIOL, 5-[[octyl-[[4-(2-oxidanylphenoxy)phenyl]methyl]amino]methyl]-2-phenoxy-phenol, ACETATE ION, ... | | 著者 | Tamhaev, R, Maveyraud, L, Chebaiki, M, Lherbet, C, Mourey, L. | | 登録日 | 2023-04-21 | | 公開日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.108 Å) | | 主引用文献 | Exploring the plasticity of the InhA substrate-binding site using new diaryl ether inhibitors.
Bioorg.Chem., 143, 2023
|
|
4ICC
 
 | | Crystal structure of human AKR1B10 complexed with NADP+ and JF0064 | | 分子名称: | 2,2',3,3',5,5',6,6'-octafluorobiphenyl-4,4'-diol, Aldo-keto reductase family 1 member B10, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | 著者 | Cousido-Siah, A, Ruiz, F.X, Mitschler, A, Porte, S, de Lera, A.R, Martin, M.J, de la Fuente, J.A, Klebe, G, Farres, J, Pares, X, Podjarny, A. | | 登録日 | 2012-12-10 | | 公開日 | 2014-02-19 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.752 Å) | | 主引用文献 | Identification of a novel polyfluorinated compound as a lead to inhibit the human enzymes aldose reductase and AKR1B10: structure determination of both ternary complexes and implications for drug design.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
3GA4
 
 | | Crystal structure of Ost6L (photoreduced form) | | 分子名称: | 1,2-ETHANEDIOL, Dolichyl-diphosphooligosaccharide-protein glycosyltransferase subunit OST6, TETRAETHYLENE GLYCOL | | 著者 | Stirnimann, C.U, Grimshaw, J.P.A, Schulz, B.L, Brozzo, M.S, Fritsch, F, Glockshuber, R, Capitani, G, Gruetter, M.G, Aebi, M. | | 登録日 | 2009-02-16 | | 公開日 | 2009-06-16 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.3 Å) | | 主引用文献 | Oxidoreductase activity of oligosaccharyltransferase subunits Ost3p and Ost6p defines site-specific glycosylation efficiency.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3G9B
 
 | | Crystal structure of reduced Ost6L | | 分子名称: | Dolichyl-diphosphooligosaccharide-protein glycosyltransferase subunit OST6 | | 著者 | Stirnimann, C.U, Grimshaw, J.P.A, Schulz, B.L, Brozzo, M.S, Fritsch, F, Glockshuber, R, Capitani, G, Gruetter, M.G, Aebi, M. | | 登録日 | 2009-02-13 | | 公開日 | 2009-06-16 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.96 Å) | | 主引用文献 | Oxidoreductase activity of oligosaccharyltransferase subunits Ost3p and Ost6p defines site-specific glycosylation efficiency.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
1B0A
 
 | | 5,10, METHYLENE-TETRAHYDROPHOLATE DEHYDROGENASE/CYCLOHYDROLASE FROM E COLI. | | 分子名称: | PROTEIN (FOLD BIFUNCTIONAL PROTEIN) | | 著者 | Shen, B.W, Dyer, D, Huang, J.-Y, D'Ari, L, Rabinowitz, J, Stoddard, B.L. | | 登録日 | 1998-11-06 | | 公開日 | 1999-06-29 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2.56 Å) | | 主引用文献 | The crystal structure of a bacterial, bifunctional 5,10 methylene-tetrahydrofolate dehydrogenase/cyclohydrolase.
Protein Sci., 8, 1999
|
|
8WCR
 
 | | Cryo-EM structure of nanodisc (PE:PS:PC) reconstituted GLIC at pH 4 in open state | | 分子名称: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, CHLORIDE ION, Proton-gated ion channel | | 著者 | Bharambe, N, Li, Z, Basak, S. | | 登録日 | 2023-09-13 | | 公開日 | 2024-04-17 | | 実験手法 | ELECTRON MICROSCOPY (2.74 Å) | | 主引用文献 | Cryo-EM structures of prokaryotic ligand-gated ion channel GLIC provide insights into gating in a lipid environment.
Nat Commun, 15, 2024
|
|
6QWQ
 
 | | Structure of gtPebB | | 分子名称: | Ferredoxin bilin reductase plastid, SULFATE ION | | 著者 | Sommerkamp, J.A, Hofmann, E. | | 登録日 | 2019-03-06 | | 公開日 | 2019-08-07 | | 最終更新日 | 2024-05-15 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Crystal structure of the first eukaryotic bilin reductaseGtPEBB reveals a flipped binding mode of dihydrobiliverdin.
J.Biol.Chem., 294, 2019
|
|
9CTX
 
 | | X-ray crystal structure of multi-drug resistant HIV-1 protease (P51) in complex with Darunavir | | 分子名称: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, GLYCEROL, Protease | | 著者 | Hayashi, H, Yedidi, R, Bulut, H, Das, D, Mitsuya, H. | | 登録日 | 2024-07-25 | | 公開日 | 2024-08-07 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | Secondary amino-acid substitutions contribute to the emergence of HIV protease inhibitor resistance as directly as primary amino-acid substitutions.
To Be Published
|
|
8SQU
 
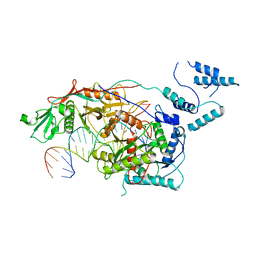 | | Monomeric MapSPARTA bound with guide RNA and target DNA hybrid | | 分子名称: | MAGNESIUM ION, TIR-APAZ, guide RNA, ... | | 著者 | Shen, Z.F, Yang, X.Y, Fu, T.M. | | 登録日 | 2023-05-04 | | 公開日 | 2023-08-23 | | 最終更新日 | 2023-09-20 | | 実験手法 | ELECTRON MICROSCOPY (3.28 Å) | | 主引用文献 | Oligomerization-mediated activation of a short prokaryotic Argonaute.
Nature, 621, 2023
|
|
