7FLX
 
 | | PanDDA analysis group deposition -- Aar2/RNaseH in complex with fragment P05G08 from the F2X-Universal Library | | 分子名称: | 2-chlorobenzohydrazide, A1 cistron-splicing factor AAR2, Pre-mRNA-splicing factor 8 | | 著者 | Barthel, T, Wollenhaupt, J, Lima, G.M.A, Wahl, M.C, Weiss, M.S. | | 登録日 | 2022-08-26 | | 公開日 | 2022-11-02 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Large-Scale Crystallographic Fragment Screening Expedites Compound Optimization and Identifies Putative Protein-Protein Interaction Sites.
J.Med.Chem., 65, 2022
|
|
7FL4
 
 | | PanDDA analysis group deposition -- Aar2/RNaseH in complex with fragment P04H06 from the F2X-Universal Library | | 分子名称: | A1 cistron-splicing factor AAR2, Pre-mRNA-splicing factor 8, [2-(methylsulfanyl)pyridin-3-yl](pyrrolidin-1-yl)methanone | | 著者 | Barthel, T, Wollenhaupt, J, Lima, G.M.A, Wahl, M.C, Weiss, M.S. | | 登録日 | 2022-08-26 | | 公開日 | 2022-11-02 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (1.51 Å) | | 主引用文献 | Large-Scale Crystallographic Fragment Screening Expedites Compound Optimization and Identifies Putative Protein-Protein Interaction Sites.
J.Med.Chem., 65, 2022
|
|
7FKS
 
 | | PanDDA analysis group deposition -- Aar2/RNaseH in complex with fragment P04F01 from the F2X-Universal Library | | 分子名称: | 4-chloro-6-(pyrrolidin-1-yl)pyrimidin-5-amine, A1 cistron-splicing factor AAR2, Pre-mRNA-splicing factor 8 | | 著者 | Barthel, T, Wollenhaupt, J, Lima, G.M.A, Wahl, M.C, Weiss, M.S. | | 登録日 | 2022-08-26 | | 公開日 | 2022-11-02 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (1.58 Å) | | 主引用文献 | Large-Scale Crystallographic Fragment Screening Expedites Compound Optimization and Identifies Putative Protein-Protein Interaction Sites.
J.Med.Chem., 65, 2022
|
|
7FKY
 
 | | PanDDA analysis group deposition -- Aar2/RNaseH in complex with fragment P04G05 from the F2X-Universal Library | | 分子名称: | 1-{[(2-methylphenyl)methyl]carbamoyl}cyclopropane-1-carboxylic acid, A1 cistron-splicing factor AAR2, Pre-mRNA-splicing factor 8 | | 著者 | Barthel, T, Wollenhaupt, J, Lima, G.M.A, Wahl, M.C, Weiss, M.S. | | 登録日 | 2022-08-26 | | 公開日 | 2022-11-02 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (1.59 Å) | | 主引用文献 | Large-Scale Crystallographic Fragment Screening Expedites Compound Optimization and Identifies Putative Protein-Protein Interaction Sites.
J.Med.Chem., 65, 2022
|
|
7FL8
 
 | | PanDDA analysis group deposition -- Aar2/RNaseH in complex with fragment P05A03 from the F2X-Universal Library | | 分子名称: | A1 cistron-splicing factor AAR2, Pre-mRNA-splicing factor 8, ethyl 4-fluoro-3-nitrobenzoate | | 著者 | Barthel, T, Wollenhaupt, J, Lima, G.M.A, Wahl, M.C, Weiss, M.S. | | 登録日 | 2022-08-26 | | 公開日 | 2022-11-02 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (1.51 Å) | | 主引用文献 | Large-Scale Crystallographic Fragment Screening Expedites Compound Optimization and Identifies Putative Protein-Protein Interaction Sites.
J.Med.Chem., 65, 2022
|
|
7FLF
 
 | | PanDDA analysis group deposition -- Aar2/RNaseH in complex with fragment P05C06 from the F2X-Universal Library | | 分子名称: | A1 cistron-splicing factor AAR2, Pre-mRNA-splicing factor 8, methyl 3-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)propanoate | | 著者 | Barthel, T, Wollenhaupt, J, Lima, G.M.A, Wahl, M.C, Weiss, M.S. | | 登録日 | 2022-08-26 | | 公開日 | 2022-11-02 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (1.54 Å) | | 主引用文献 | Large-Scale Crystallographic Fragment Screening Expedites Compound Optimization and Identifies Putative Protein-Protein Interaction Sites.
J.Med.Chem., 65, 2022
|
|
7FL3
 
 | | PanDDA analysis group deposition -- Aar2/RNaseH in complex with fragment P04H06 from the F2X-Universal Library | | 分子名称: | A1 cistron-splicing factor AAR2, Pre-mRNA-splicing factor 8, [2-(methylsulfanyl)pyridin-3-yl](pyrrolidin-1-yl)methanone | | 著者 | Barthel, T, Wollenhaupt, J, Lima, G.M.A, Wahl, M.C, Weiss, M.S. | | 登録日 | 2022-08-26 | | 公開日 | 2022-11-02 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (1.51 Å) | | 主引用文献 | Large-Scale Crystallographic Fragment Screening Expedites Compound Optimization and Identifies Putative Protein-Protein Interaction Sites.
J.Med.Chem., 65, 2022
|
|
7FM4
 
 | | PanDDA analysis group deposition -- Aar2/RNaseH in complex with fragment P06A01 from the F2X-Universal Library | | 分子名称: | (4Z)-6-fluoro-4-(methoxyimino)-3,4-dihydro-1-benzothiopyran-1,1(2H)-dione, A1 cistron-splicing factor AAR2, Pre-mRNA-splicing factor 8 | | 著者 | Barthel, T, Wollenhaupt, J, Lima, G.M.A, Wahl, M.C, Weiss, M.S. | | 登録日 | 2022-08-26 | | 公開日 | 2022-11-02 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (1.62 Å) | | 主引用文献 | Large-Scale Crystallographic Fragment Screening Expedites Compound Optimization and Identifies Putative Protein-Protein Interaction Sites.
J.Med.Chem., 65, 2022
|
|
7FLK
 
 | | PanDDA analysis group deposition -- Aar2/RNaseH in complex with fragment P05E08 from the F2X-Universal Library | | 分子名称: | (5R,8aS)-8a-phenylhexahydropyrrolo[1,2-a]pyrimidin-6(2H)-one, A1 cistron-splicing factor AAR2, Pre-mRNA-splicing factor 8 | | 著者 | Barthel, T, Wollenhaupt, J, Lima, G.M.A, Wahl, M.C, Weiss, M.S. | | 登録日 | 2022-08-26 | | 公開日 | 2022-11-02 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (1.62 Å) | | 主引用文献 | Large-Scale Crystallographic Fragment Screening Expedites Compound Optimization and Identifies Putative Protein-Protein Interaction Sites.
J.Med.Chem., 65, 2022
|
|
7FLR
 
 | | PanDDA analysis group deposition -- Aar2/RNaseH in complex with fragment P05G01 from the F2X-Universal Library | | 分子名称: | 2-(aminomethyl)-N,N-dimethyl-2,3-dihydro-1H-inden-2-amine, A1 cistron-splicing factor AAR2, Pre-mRNA-splicing factor 8 | | 著者 | Barthel, T, Wollenhaupt, J, Lima, G.M.A, Wahl, M.C, Weiss, M.S. | | 登録日 | 2022-08-26 | | 公開日 | 2022-11-02 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (1.63 Å) | | 主引用文献 | Large-Scale Crystallographic Fragment Screening Expedites Compound Optimization and Identifies Putative Protein-Protein Interaction Sites.
J.Med.Chem., 65, 2022
|
|
7FLY
 
 | | PanDDA analysis group deposition -- Aar2/RNaseH in complex with fragment P05G10 from the F2X-Universal Library | | 分子名称: | A1 cistron-splicing factor AAR2, Pre-mRNA-splicing factor 8, piperidine-4-carboxamide | | 著者 | Barthel, T, Wollenhaupt, J, Lima, G.M.A, Wahl, M.C, Weiss, M.S. | | 登録日 | 2022-08-26 | | 公開日 | 2022-11-02 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | Large-Scale Crystallographic Fragment Screening Expedites Compound Optimization and Identifies Putative Protein-Protein Interaction Sites.
J.Med.Chem., 65, 2022
|
|
7FM8
 
 | | PanDDA analysis group deposition -- Aar2/RNaseH in complex with fragment P06A09 from the F2X-Universal Library | | 分子名称: | (5S,6Z)-6-imino-1,3-dimethyl-5-propyl-1,3-diazinane-2,4-dione, A1 cistron-splicing factor AAR2, Pre-mRNA-splicing factor 8 | | 著者 | Barthel, T, Wollenhaupt, J, Lima, G.M.A, Wahl, M.C, Weiss, M.S. | | 登録日 | 2022-08-26 | | 公開日 | 2022-11-02 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | Large-Scale Crystallographic Fragment Screening Expedites Compound Optimization and Identifies Putative Protein-Protein Interaction Sites.
J.Med.Chem., 65, 2022
|
|
7FO9
 
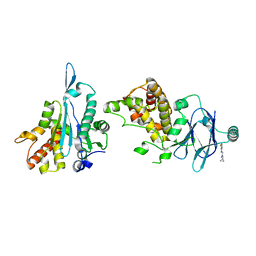 | | PanDDA analysis group deposition -- Aar2/RNaseH in complex with fragment P07H07 from the F2X-Universal Library | | 分子名称: | A1 cistron-splicing factor AAR2, N-cyclopropyl-2-(3-fluorophenoxy)acetamide, Pre-mRNA-splicing factor 8 | | 著者 | Barthel, T, Wollenhaupt, J, Lima, G.M.A, Wahl, M.C, Weiss, M.S. | | 登録日 | 2022-08-26 | | 公開日 | 2022-11-02 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (1.67 Å) | | 主引用文献 | Large-Scale Crystallographic Fragment Screening Expedites Compound Optimization and Identifies Putative Protein-Protein Interaction Sites.
J.Med.Chem., 65, 2022
|
|
7FKB
 
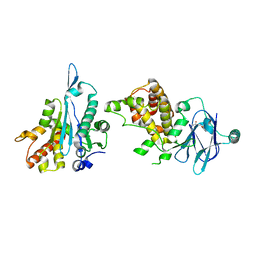 | | PanDDA analysis group deposition -- Aar2/RNaseH in complex with fragment P04C07 from the F2X-Universal Library | | 分子名称: | A1 cistron-splicing factor AAR2, N-[(2-methoxyphenyl)methyl]urea, Pre-mRNA-splicing factor 8 | | 著者 | Barthel, T, Wollenhaupt, J, Lima, G.M.A, Wahl, M.C, Weiss, M.S. | | 登録日 | 2022-08-26 | | 公開日 | 2022-11-02 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (1.68 Å) | | 主引用文献 | Large-Scale Crystallographic Fragment Screening Expedites Compound Optimization and Identifies Putative Protein-Protein Interaction Sites.
J.Med.Chem., 65, 2022
|
|
7FM5
 
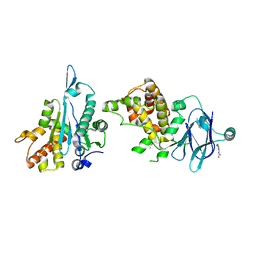 | | PanDDA analysis group deposition -- Aar2/RNaseH in complex with fragment P06A02 from the F2X-Universal Library | | 分子名称: | A1 cistron-splicing factor AAR2, N-(2-amino-5-chlorophenyl)acetamide, Pre-mRNA-splicing factor 8 | | 著者 | Barthel, T, Wollenhaupt, J, Lima, G.M.A, Wahl, M.C, Weiss, M.S. | | 登録日 | 2022-08-26 | | 公開日 | 2022-11-02 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (1.56 Å) | | 主引用文献 | Large-Scale Crystallographic Fragment Screening Expedites Compound Optimization and Identifies Putative Protein-Protein Interaction Sites.
J.Med.Chem., 65, 2022
|
|
7FMD
 
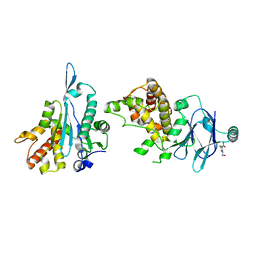 | | PanDDA analysis group deposition -- Aar2/RNaseH in complex with fragment P06B12 from the F2X-Universal Library | | 分子名称: | 2-(4-chloro-2-nitroanilino)ethan-1-ol, A1 cistron-splicing factor AAR2, Pre-mRNA-splicing factor 8 | | 著者 | Barthel, T, Wollenhaupt, J, Lima, G.M.A, Wahl, M.C, Weiss, M.S. | | 登録日 | 2022-08-26 | | 公開日 | 2022-11-02 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (1.68 Å) | | 主引用文献 | Large-Scale Crystallographic Fragment Screening Expedites Compound Optimization and Identifies Putative Protein-Protein Interaction Sites.
J.Med.Chem., 65, 2022
|
|
7FMJ
 
 | | PanDDA analysis group deposition -- Aar2/RNaseH in complex with fragment P06C12 from the F2X-Universal Library | | 分子名称: | (2E)-3-[4-(2-oxopyrrolidin-1-yl)phenyl]prop-2-enoic acid, A1 cistron-splicing factor AAR2, Pre-mRNA-splicing factor 8 | | 著者 | Barthel, T, Wollenhaupt, J, Lima, G.M.A, Wahl, M.C, Weiss, M.S. | | 登録日 | 2022-08-26 | | 公開日 | 2022-11-02 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Large-Scale Crystallographic Fragment Screening Expedites Compound Optimization and Identifies Putative Protein-Protein Interaction Sites.
J.Med.Chem., 65, 2022
|
|
7FMQ
 
 | | PanDDA analysis group deposition -- Aar2/RNaseH in complex with fragment P06E04 from the F2X-Universal Library | | 分子名称: | 3-phenoxypropanoic acid, A1 cistron-splicing factor AAR2, Pre-mRNA-splicing factor 8 | | 著者 | Barthel, T, Wollenhaupt, J, Lima, G.M.A, Wahl, M.C, Weiss, M.S. | | 登録日 | 2022-08-26 | | 公開日 | 2022-11-02 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (1.58 Å) | | 主引用文献 | Large-Scale Crystallographic Fragment Screening Expedites Compound Optimization and Identifies Putative Protein-Protein Interaction Sites.
J.Med.Chem., 65, 2022
|
|
7FKI
 
 | | PanDDA analysis group deposition -- Aar2/RNaseH in complex with fragment P04D08 from the F2X-Universal Library | | 分子名称: | (2R)-(3,4-difluorophenyl)(hydroxy)acetic acid, A1 cistron-splicing factor AAR2, Pre-mRNA-splicing factor 8 | | 著者 | Barthel, T, Wollenhaupt, J, Lima, G.M.A, Wahl, M.C, Weiss, M.S. | | 登録日 | 2022-08-26 | | 公開日 | 2022-11-02 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (1.47 Å) | | 主引用文献 | Large-Scale Crystallographic Fragment Screening Expedites Compound Optimization and Identifies Putative Protein-Protein Interaction Sites.
J.Med.Chem., 65, 2022
|
|
7FKN
 
 | | PanDDA analysis group deposition -- Aar2/RNaseH in complex with fragment P04E05 from the F2X-Universal Library | | 分子名称: | A1 cistron-splicing factor AAR2, P-HYDROXYACETOPHENONE, Pre-mRNA-splicing factor 8 | | 著者 | Barthel, T, Wollenhaupt, J, Lima, G.M.A, Wahl, M.C, Weiss, M.S. | | 登録日 | 2022-08-26 | | 公開日 | 2022-11-02 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | Large-Scale Crystallographic Fragment Screening Expedites Compound Optimization and Identifies Putative Protein-Protein Interaction Sites.
J.Med.Chem., 65, 2022
|
|
7FKT
 
 | | PanDDA analysis group deposition -- Aar2/RNaseH in complex with fragment P04F02 from the F2X-Universal Library | | 分子名称: | A1 cistron-splicing factor AAR2, N~2~-(3-chlorophenyl)-N~2~-(methanesulfonyl)-N-methylglycinamide, Pre-mRNA-splicing factor 8 | | 著者 | Barthel, T, Wollenhaupt, J, Lima, G.M.A, Wahl, M.C, Weiss, M.S. | | 登録日 | 2022-08-26 | | 公開日 | 2022-11-02 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (1.44 Å) | | 主引用文献 | Large-Scale Crystallographic Fragment Screening Expedites Compound Optimization and Identifies Putative Protein-Protein Interaction Sites.
J.Med.Chem., 65, 2022
|
|
7FKX
 
 | | PanDDA analysis group deposition -- Aar2/RNaseH in complex with fragment P04G02 from the F2X-Universal Library | | 分子名称: | 1-(4-chlorophenyl)cyclobutane-1-carboxylic acid, A1 cistron-splicing factor AAR2, Pre-mRNA-splicing factor 8 | | 著者 | Barthel, T, Wollenhaupt, J, Lima, G.M.A, Wahl, M.C, Weiss, M.S. | | 登録日 | 2022-08-26 | | 公開日 | 2022-11-02 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Large-Scale Crystallographic Fragment Screening Expedites Compound Optimization and Identifies Putative Protein-Protein Interaction Sites.
J.Med.Chem., 65, 2022
|
|
7FMX
 
 | | PanDDA analysis group deposition -- Aar2/RNaseH in complex with fragment P06F06 from the F2X-Universal Library | | 分子名称: | 3-phenyl-1,2-oxazole-5-carboxylic acid, A1 cistron-splicing factor AAR2, Pre-mRNA-splicing factor 8 | | 著者 | Barthel, T, Wollenhaupt, J, Lima, G.M.A, Wahl, M.C, Weiss, M.S. | | 登録日 | 2022-08-26 | | 公開日 | 2022-11-02 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (1.51 Å) | | 主引用文献 | Large-Scale Crystallographic Fragment Screening Expedites Compound Optimization and Identifies Putative Protein-Protein Interaction Sites.
J.Med.Chem., 65, 2022
|
|
7FN1
 
 | | PanDDA analysis group deposition -- Aar2/RNaseH in complex with fragment P06G06 from the F2X-Universal Library | | 分子名称: | 4-amino-3-methyl-N-(propan-2-yl)benzamide, A1 cistron-splicing factor AAR2, Pre-mRNA-splicing factor 8 | | 著者 | Barthel, T, Wollenhaupt, J, Lima, G.M.A, Wahl, M.C, Weiss, M.S. | | 登録日 | 2022-08-26 | | 公開日 | 2022-11-02 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (1.44 Å) | | 主引用文献 | Large-Scale Crystallographic Fragment Screening Expedites Compound Optimization and Identifies Putative Protein-Protein Interaction Sites.
J.Med.Chem., 65, 2022
|
|
7FNC
 
 | | PanDDA analysis group deposition -- Aar2/RNaseH in complex with fragment P07B01 from the F2X-Universal Library | | 分子名称: | A1 cistron-splicing factor AAR2, N-{2-[(4R)-8-methylimidazo[1,2-a]pyridin-2-yl]ethyl}acetamide, Pre-mRNA-splicing factor 8 | | 著者 | Barthel, T, Wollenhaupt, J, Lima, G.M.A, Wahl, M.C, Weiss, M.S. | | 登録日 | 2022-08-26 | | 公開日 | 2022-11-02 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | Large-Scale Crystallographic Fragment Screening Expedites Compound Optimization and Identifies Putative Protein-Protein Interaction Sites.
J.Med.Chem., 65, 2022
|
|
