4BF4
 
 | |
7JXB
 
 | | The crystal structure of 4-(3'-methoxyphenyl)benzoic acid-bound CYP199A4 | | 分子名称: | 3'-methoxy[1,1'-biphenyl]-4-carboxylic acid, CHLORIDE ION, Cytochrome P450, ... | | 著者 | Doherty, D.Z, Bell, S.G, Bruning, J. | | 登録日 | 2020-08-27 | | 公開日 | 2022-03-09 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.655 Å) | | 主引用文献 | Enabling Aromatic Hydroxylation in a Cytochrome P450 Monooxygenase Enzyme through Protein Engineering.
Chemistry, 28, 2022
|
|
7KHW
 
 | |
5U6U
 
 | | The crystal structure of 4-ethylthiobenzoate-bound CYP199A4 | | 分子名称: | 4-(ethylsulfanyl)benzoic acid, CHLORIDE ION, Cytochrome P450, ... | | 著者 | Coleman, T, Bruning, J.B, Bell, S.G. | | 登録日 | 2016-12-08 | | 公開日 | 2018-06-13 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.786 Å) | | 主引用文献 | Cytochrome P450 CYP199A4 from Rhodopseudomonas palustris Catalyzes Heteroatom Dealkylations, Sulfoxidation, and Amide and Cyclic Hemiacetal Formation
Acs Catalysis, 8, 2018
|
|
5U6T
 
 | | The crystal structure of 4-ethoxybenzoate-bound CYP199A4 | | 分子名称: | 4-ethoxybenzoic acid, CHLORIDE ION, Cytochrome P450, ... | | 著者 | Coleman, T, Bruning, J.B, Bell, S.G. | | 登録日 | 2016-12-08 | | 公開日 | 2018-06-13 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.935 Å) | | 主引用文献 | Cytochrome P450 CYP199A4 from Rhodopseudomonas palustris Catalyzes Heteroatom Dealkylations, Sulfoxidation, and Amide and Cyclic Hemiacetal Formation
Acs Catalysis, 8, 2018
|
|
5T0Q
 
 | | Crystal structure of the Myc3 N-terminal domain [44-242] in complex with JAZ10 Jas domain [166-192] from arabidopsis | | 分子名称: | Protein TIFY 9, Transcription factor MYC3 | | 著者 | Ke, J, Zhang, F, Brunzelle, J.S, He, S.Y, Xu, H.E, Melcher, K. | | 登録日 | 2016-08-16 | | 公開日 | 2017-01-25 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.15 Å) | | 主引用文献 | Structural insights into alternative splicing-mediated desensitization of jasmonate signaling.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5T0F
 
 | | Crystal structure of the Myc3 N-terminal domain [44-242] in complex with JAZ10 CMID domain [16-58] from arabidopsis | | 分子名称: | Protein TIFY 9, Transcription factor MYC3 | | 著者 | Ke, J, Zhang, F, Brunzelle, J.S, He, S.Y, Xu, H.E, Melcher, K. | | 登録日 | 2016-08-16 | | 公開日 | 2017-01-25 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Structural insights into alternative splicing-mediated desensitization of jasmonate signaling.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
8QLE
 
 | | Crystal structure of the light-driven sodium pump ErNaR in the monomeric form at pH 4.6 | | 分子名称: | Bacteriorhodopsin-like protein, EICOSANE, OLEIC ACID | | 著者 | Kovalev, K, Podoliak, E, Lamm, G.H.U, Astashkin, R, Bourenkov, G. | | 登録日 | 2023-09-19 | | 公開日 | 2024-04-24 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | A subgroup of light-driven sodium pumps with an additional Schiff base counterion.
Nat Commun, 15, 2024
|
|
8QLF
 
 | | Crystal structure of the light-driven sodium pump ErNaR in the monomeric form at pH 8.8 | | 分子名称: | Bacteriorhodopsin-like protein, EICOSANE, OLEIC ACID | | 著者 | Kovalev, K, Podoliak, E, Lamm, G.H.U, Astashkin, R, Bourenkov, G. | | 登録日 | 2023-09-19 | | 公開日 | 2024-04-24 | | 実験手法 | X-RAY DIFFRACTION (1.71 Å) | | 主引用文献 | A subgroup of light-driven sodium pumps with an additional Schiff base counterion.
Nat Commun, 15, 2024
|
|
8QQZ
 
 | | Cryo-EM structure of the light-driven sodium pump ErNaR in the pentameric form at pH 8.0 | | 分子名称: | Bacteriorhodopsin-like protein, DODECYL-BETA-D-MALTOSIDE, EICOSANE | | 著者 | Kovalev, K, Podoliak, E, Lamm, G.H.U, Marin, E, Stetsenko, A, Guskov, A. | | 登録日 | 2023-10-06 | | 公開日 | 2024-04-24 | | 実験手法 | ELECTRON MICROSCOPY (2.63 Å) | | 主引用文献 | A subgroup of light-driven sodium pumps with an additional Schiff base counterion.
Nat Commun, 15, 2024
|
|
8QR0
 
 | | Cryo-EM structure of the light-driven sodium pump ErNaR in the pentameric form at pH 4.3 | | 分子名称: | Bacteriorhodopsin-like protein, DODECYL-BETA-D-MALTOSIDE, EICOSANE | | 著者 | Kovalev, K, Podoliak, E, Lamm, G.H.U, Marin, E, Stetsenko, A, Guskov, A. | | 登録日 | 2023-10-06 | | 公開日 | 2024-04-24 | | 実験手法 | ELECTRON MICROSCOPY (2.5 Å) | | 主引用文献 | A subgroup of light-driven sodium pumps with an additional Schiff base counterion.
Nat Commun, 15, 2024
|
|
8Q5J
 
 | |
7JXG
 
 | | Structural model for Fe-containing human acireductone dioxygenase | | 分子名称: | 1,2-dihydroxy-3-keto-5-methylthiopentene dioxygenase, FE (II) ION | | 著者 | Pochapsky, T.C, Liu, X, Deshpande, A, Ringe, D, Garber, A, Ryan, J. | | 登録日 | 2020-08-27 | | 公開日 | 2020-11-18 | | 最終更新日 | 2024-05-01 | | 実験手法 | SOLUTION NMR | | 主引用文献 | A Model for the Solution Structure of Human Fe(II)-Bound Acireductone Dioxygenase and Interactions with the Regulatory Domain of Matrix Metalloproteinase I (MMP-I).
Biochemistry, 59, 2020
|
|
7L1E
 
 | | The Crystal Structure of Bromide-Bound GtACR1 | | 分子名称: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Anion channelrhodopsin-1, BROMIDE ION, ... | | 著者 | Li, H, Huang, C.Y, Wang, M, Spudich, J.L, Zheng, L. | | 登録日 | 2020-12-14 | | 公開日 | 2021-05-26 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (3.2 Å) | | 主引用文献 | The crystal structure of bromide-bound Gt ACR1 reveals a pre-activated state in the transmembrane anion tunnel.
Elife, 10, 2021
|
|
4CPP
 
 | | CRYSTAL STRUCTURES OF CYTOCHROME P450-CAM COMPLEXED WITH CAMPHANE, THIOCAMPHOR, AND ADAMANTANE: FACTORS CONTROLLING P450 SUBSTRATE HYDROXYLATION | | 分子名称: | ADAMANTANE, CYTOCHROME P450-CAM, PROTOPORPHYRIN IX CONTAINING FE | | 著者 | Raag, R, Poulos, T.L. | | 登録日 | 1990-05-18 | | 公開日 | 1991-07-15 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.11 Å) | | 主引用文献 | Crystal structures of cytochrome P-450CAM complexed with camphane, thiocamphor, and adamantane: factors controlling P-450 substrate hydroxylation.
Biochemistry, 30, 1991
|
|
4E2P
 
 | | Crystal Structure of a Post-tailoring Hydroxylase (HmtN) Involved in the Himastatin Biosynthesis | | 分子名称: | Cytochrome P450 107B1 (P450CVIIB1), MAGNESIUM ION, PROTOPORPHYRIN IX CONTAINING FE | | 著者 | Zhang, H.D, Chen, J, Wang, H, Huang, L, Zhang, H.J. | | 登録日 | 2012-03-09 | | 公開日 | 2013-03-13 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.36 Å) | | 主引用文献 | Crystal Structure of a Post-tailoring Hydroxylase (HmtN) Involved in the Himastatin Biosynthesis
To be Published
|
|
4EGO
 
 | | The X-ray crystal structure of CYP199A4 in complex with indole-6-carboxylic acid | | 分子名称: | 1H-indole-6-carboxylic acid, CHLORIDE ION, Cytochrome P450, ... | | 著者 | Zhou, W, Bell, S.G, Yang, W, Zhou, R.M, Tan, A.B.H, Wong, L.-L. | | 登録日 | 2012-03-31 | | 公開日 | 2013-02-20 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.76 Å) | | 主引用文献 | Investigation of the substrate range of CYP199A4: modification of the partition between hydroxylation and desaturation activities by substrate and protein engineering
Chemistry, 18, 2012
|
|
4EGM
 
 | | The X-ray crystal structure of CYP199A4 in complex with 4-ethylbenzoic acid | | 分子名称: | 4-ethylbenzoic acid, CHLORIDE ION, Cytochrome P450, ... | | 著者 | Zhou, W, Bell, S.G, Yang, W, Zhou, R.M, Tan, A.B.H, Wong, L.-L. | | 登録日 | 2012-03-31 | | 公開日 | 2013-02-20 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.91 Å) | | 主引用文献 | Investigation of the substrate range of CYP199A4: modification of the partition between hydroxylation and desaturation activities by substrate and protein engineering
Chemistry, 18, 2012
|
|
7KCS
 
 | | The crystal structure of 4-vinylbenzoate-bound wild-type CYP199A4 | | 分子名称: | 4-ethenylbenzoic acid, CHLORIDE ION, Cytochrome P450, ... | | 著者 | Coleman, T, Bruning, J.B, Bell, S.G. | | 登録日 | 2020-10-07 | | 公開日 | 2021-05-19 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.773 Å) | | 主引用文献 | Understanding the Mechanistic Requirements for Efficient and Stereoselective Alkene Epoxidation by a Cytochrome P450 Enzyme
Acs Catalysis, 11, 2021
|
|
4DO1
 
 | | The crystal structures of 4-methoxybenzoate bound CYP199A4 | | 分子名称: | 4-METHOXYBENZOIC ACID, CHLORIDE ION, Cytochrome P450, ... | | 著者 | Zhou, W, Bell, S.G, Yang, W, Tan, A.B.H, Zhou, R, Johnson, E.O.D, Zhang, A, Rao, Z, Wong, L.-L. | | 登録日 | 2012-02-09 | | 公開日 | 2012-08-15 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | The crystal structures of 4-methoxybenzoate bound CYP199A2 and CYP199A4: structural changes on substrate binding and the identification of an anion binding site
Dalton Trans, 41, 2012
|
|
4DNJ
 
 | | The crystal structures of 4-methoxybenzoate bound CYP199A2 | | 分子名称: | 4-METHOXYBENZOIC ACID, CHLORIDE ION, PROTOPORPHYRIN IX CONTAINING FE, ... | | 著者 | Zhou, W, Bell, S.G, Yang, W, Tan, A.B.H, Zhou, R, Johnson, E.O.D, Zhang, A, Rao, Z, Wong, L.-L. | | 登録日 | 2012-02-08 | | 公開日 | 2012-08-15 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | The crystal structures of 4-methoxybenzoate bound CYP199A2 and CYP199A4: structural changes on substrate binding and the identification of an anion binding site
Dalton Trans, 41, 2012
|
|
4DNZ
 
 | | The crystal structures of CYP199A4 | | 分子名称: | CHLORIDE ION, Cytochrome P450, GLYCEROL, ... | | 著者 | Zhou, W, Bell, S.G, Yang, W, Tan, A.B.H, Zhou, R, Johnson, E.O.D, Zhang, A, Rao, Z, Wong, L.-L. | | 登録日 | 2012-02-09 | | 公開日 | 2012-08-15 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | The crystal structures of 4-methoxybenzoate bound CYP199A2 and CYP199A4: structural changes on substrate binding and the identification of an anion binding site
Dalton Trans, 41, 2012
|
|
8SPC
 
 | |
4DAJ
 
 | | Structure of the M3 Muscarinic Acetylcholine Receptor | | 分子名称: | (1R,2R,4S,5S,7S)-7-{[hydroxy(dithiophen-2-yl)acetyl]oxy}-9,9-dimethyl-3-oxa-9-azoniatricyclo[3.3.1.0~2,4~]nonane, Muscarinic acetylcholine receptor M3, Lysozyme, ... | | 著者 | Kruse, A.C, Hu, J, Pan, A.C, Arlow, D.H, Rosenbaum, D.M, Rosemond, E, Green, H.F, Liu, T, Chae, P.S, Dror, R.O, Shaw, D.E, Weis, W.I, Wess, J, Kobilka, B. | | 登録日 | 2012-01-12 | | 公開日 | 2012-02-22 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (3.4 Å) | | 主引用文献 | Structure and dynamics of the M3 muscarinic acetylcholine receptor.
Nature, 482, 2012
|
|
5UK6
 
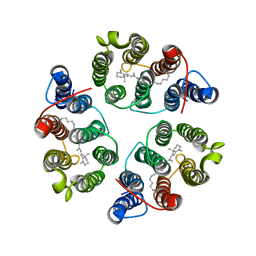 | | Structure of Anabaena Sensory Rhodopsin Determined by Solid State NMR Spectroscopy and DEER | | 分子名称: | Bacteriorhodopsin | | 著者 | Milikisiyants, S, Wang, S, Munro, R.A, Donohue, M, Ward, M.E, Brown, L.S, Smirnova, T.I, Ladizhansky, V, Smirnov, A.I. | | 登録日 | 2017-01-20 | | 公開日 | 2017-05-31 | | 最終更新日 | 2020-01-08 | | 実験手法 | SOLID-STATE NMR | | 主引用文献 | Oligomeric Structure of Anabaena Sensory Rhodopsin in a Lipid Bilayer Environment by Combining Solid-State NMR and Long-range DEER Constraints.
J. Mol. Biol., 429, 2017
|
|
