1SZA
 
 | |
1GUF
 
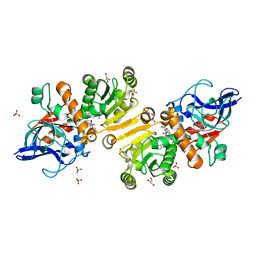 | | Enoyl thioester reductase from Candida tropicalis | | 分子名称: | ENOYL-[ACYL-CARRIER-PROTEIN] REDUCTASE [NADPH, B-SPECIFIC] 1, MITOCHONDRIAL, ... | | 著者 | Airenne, T.T, Torkko, J.M, Van Der Plas, S, Sormunen, R.T, Kastaniotis, A.J, Wierenga, R.K, Hiltunen, J.K. | | 登録日 | 2002-01-25 | | 公開日 | 2003-03-13 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | Structure-Function Analysis of Enoyl Thioester Reductase Involved in Mitochondrial Maintenance
J.Mol.Biol., 327, 2003
|
|
1CXN
 
 | | REFINED THREE-DIMENSIONAL SOLUTION STRUCTURE OF A SNAKE CARDIOTOXIN: ANALYSIS OF THE SIDE-CHAIN ORGANISATION SUGGESTS THE EXISTENCE OF A POSSIBLE PHOSPHOLIPID BINDING SITE | | 分子名称: | CARDIOTOXIN GAMMA | | 著者 | Gilquin, B, Roumestand, C, Zinn-Justin, S, Menez, A, Toma, F. | | 登録日 | 1994-07-08 | | 公開日 | 1994-12-20 | | 最終更新日 | 2022-02-16 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Refined three-dimensional solution structure of a snake cardiotoxin: analysis of the side-chain organization suggests the existence of a possible phospholipid binding site.
Biopolymers, 33, 1993
|
|
1D7H
 
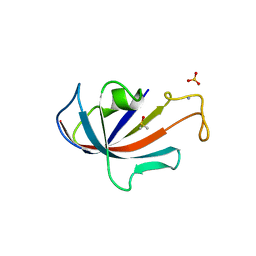 | | FKBP COMPLEXED WITH DMSO | | 分子名称: | AMMONIUM ION, DIMETHYL SULFOXIDE, PROTEIN (FK506-BINDING PROTEIN), ... | | 著者 | Burkhard, P, Taylor, P, Walkinshaw, M.D. | | 登録日 | 1999-10-18 | | 公開日 | 1999-10-21 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | X-ray structures of small ligand-FKBP complexes provide an estimate for hydrophobic interaction energies.
J.Mol.Biol., 295, 2000
|
|
1H79
 
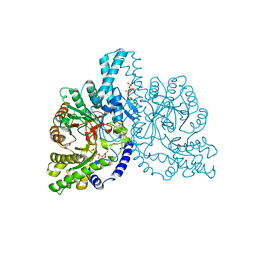 | | STRUCTURAL BASIS FOR ALLOSTERIC SUBSTRATE SPECIFICITY REGULATION IN CLASS III RIBONUCLEOTIDE REDUCTASES: NRDD IN COMPLEX WITH DTTP | | 分子名称: | ANAEROBIC RIBONUCLEOTIDE-TRIPHOSPHATE REDUCTASE LARGE CHAIN, FE (II) ION, MAGNESIUM ION, ... | | 著者 | Larsson, K.-M, Andersson, J, Sjoeberg, B.-M, Nordlund, P, Logan, D.T. | | 登録日 | 2001-07-04 | | 公開日 | 2002-03-28 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Structural Basis for Allosteric Substrate Specificty Regulation in Anaerobic Ribonucleotide Reductase
Structure, 9, 2001
|
|
1XLI
 
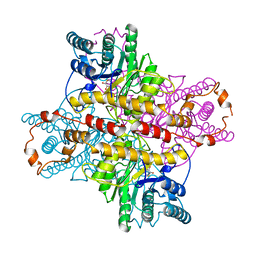 | | MECHANISM FOR ALDOSE-KETOSE INTERCONVERSION BY D-XYLOSE ISOMERASE INVOLVING RING OPENING FOLLOWED BY A 1,2-HYDRIDE SHIFT | | 分子名称: | 5-thio-alpha-D-glucopyranose, D-XYLOSE ISOMERASE, MANGANESE (II) ION | | 著者 | Collyer, C.A, Henrick, K, Blow, D.M. | | 登録日 | 1991-10-09 | | 公開日 | 1993-07-15 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Mechanism for aldose-ketose interconversion by D-xylose isomerase involving ring opening followed by a 1,2-hydride shift.
J.Mol.Biol., 212, 1990
|
|
1LC1
 
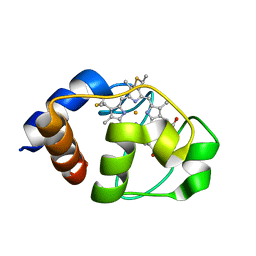 | |
1H7R
 
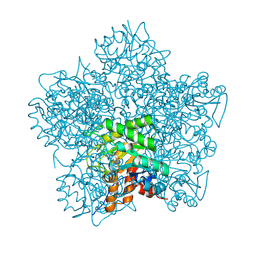 | | SCHIFF-BASE COMPLEX OF YEAST 5-AMINOLAEVULINIC ACID DEHYDRATASE WITH SUCCINYLACETONE AT 2.0 A RESOLUTION. | | 分子名称: | 4,6-DIOXOHEPTANOIC ACID, 5-AMINOLAEVULINIC ACID DEHYDRATASE, ZINC ION | | 著者 | Erskine, P.T, Newbold, R, Brindley, A.A, Wood, S.P, Shoolingin-Jordan, P.M, Warren, M.J, Cooper, J.B. | | 登録日 | 2001-07-09 | | 公開日 | 2001-07-12 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | The X-Ray Structure of Yeast 5-Aminolaevulinic Acid Dehydratase Complexed with Substrate and Three Inhibitors
J.Mol.Biol., 312, 2001
|
|
1TCU
 
 | | Crystal Structure of the Purine Nucleoside Phosphorylase from Schistosoma mansoni in complex with phosphate and acetate | | 分子名称: | ACETATE ION, DIMETHYL SULFOXIDE, PHOSPHATE ION, ... | | 著者 | Pereira, H.D, Franco, G.R, Cleasby, A, Garratt, R.C. | | 登録日 | 2004-05-21 | | 公開日 | 2005-05-24 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structures for the Potential Drug Target Purine Nucleoside Phosphorylase from Schistosoma mansoni Causal Agent of Schistosomiasis.
J.Mol.Biol., 353, 2005
|
|
1H7N
 
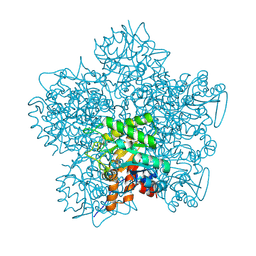 | | SCHIFF-BASE COMPLEX OF YEAST 5-AMINOLAEVULINIC ACID DEHYDRATASE WITH LAEVULINIC ACID AT 1.6 A RESOLUTION | | 分子名称: | 5-AMINOLAEVULINIC ACID DEHYDRATASE, LAEVULINIC ACID, ZINC ION | | 著者 | Erskine, P.T, Newbold, R, Brindley, A.A, Wood, S.P, Shoolingin-Jordan, P.M, Warren, M.J, Cooper, J.B. | | 登録日 | 2001-07-09 | | 公開日 | 2001-07-10 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | The X-Ray Structure of Yeast 5-Aminolaevulinic Acid Dehydratase Complexed with Substrate and Three Inhibitors
J.Mol.Biol., 312, 2001
|
|
2IL8
 
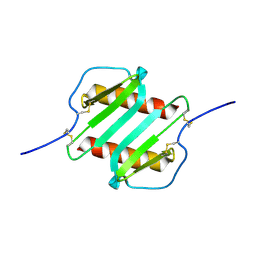 | |
1TKT
 
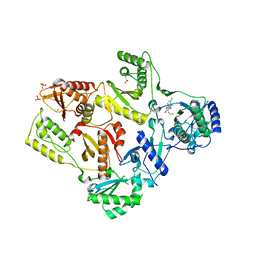 | | CRYSTAL STRUCTURE OF HIV-1 REVERSE TRANSCRIPTASE IN COMPLEX WITH GW426318 | | 分子名称: | 6-CHLORO-4-(CYCLOHEXYLOXY)-3-PROPYLQUINOLIN-2(1H)-ONE, MAGNESIUM ION, PHOSPHATE ION, ... | | 著者 | Hopkins, A.L, Ren, J, Stuart, D.I, Stammers, D.K. | | 登録日 | 2004-06-09 | | 公開日 | 2004-12-07 | | 最終更新日 | 2018-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Design of non-nucleoside inhibitors of HIV-1 reverse transcriptase with improved drug resistance properties. 1.
J.Med.Chem., 47, 2004
|
|
1H7A
 
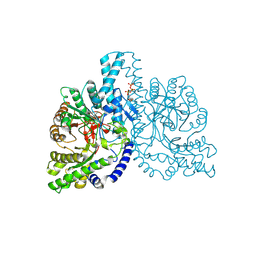 | | Structural basis for allosteric substrate specificity regulation in class III ribonucleotide reductases: NRDD in complex with dATP | | 分子名称: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, ANAEROBIC RIBONUCLEOTIDE-TRIPHOSPHATE REDUCTASE LARGE CHAIN, FE (II) ION, ... | | 著者 | Larsson, K.-M, Andersson, J, Sjoeberg, B.-M, Nordlund, P, Logan, D.T. | | 登録日 | 2001-07-04 | | 公開日 | 2002-03-28 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2.75 Å) | | 主引用文献 | Structural Basis for Allosteric Substrate Specificty Regulation in Anaerobic Ribonucleotide Reductase
Structure, 9, 2001
|
|
2J8D
 
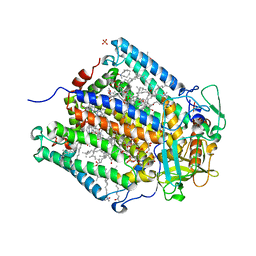 | | X-ray high resolution structure of the photosynthetic reaction center from Rb. sphaeroides at pH 8 in the charge-separated state | | 分子名称: | BACTERIOCHLOROPHYLL A, BACTERIOPHEOPHYTIN A, CARDIOLIPIN, ... | | 著者 | Koepke, J, Diehm, R, Fritzsch, G. | | 登録日 | 2006-10-24 | | 公開日 | 2007-07-03 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2.07 Å) | | 主引用文献 | Ph Modulates the Quinone Position in the Photosynthetic Reaction Center from Rhodobacter Sphaeroides in the Neutral and Charge Separated States.
J.Mol.Biol., 371, 2007
|
|
1UZW
 
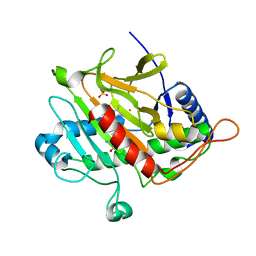 | | ISOPENICILLIN N SYNTHASE WITH L-D-(A-AMINOADIPOYL)-L-CYSTEINYL-D-ISODEHYDROVALINE | | 分子名称: | D-(L-A-AMINOADIPOYL)-L-CYSTEINYL-D-ISODEHYDROVALINE, FE (II) ION, ISOPENICILLIN N SYNTHETASE, ... | | 著者 | Grummitt, A.R, Rutledge, P.J, Clifton, I.J, Baldwin, J.E. | | 登録日 | 2004-03-17 | | 公開日 | 2004-06-10 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.3 Å) | | 主引用文献 | Active Site Mediated Elimination of Hydrogen Fluoride from a Fluorinated Substrate Analogue by Isopenicillin N Synthase
Biochem.J., 382, 2004
|
|
1D7J
 
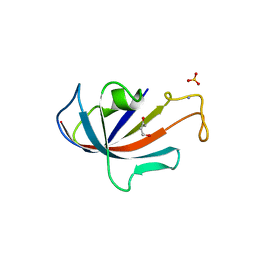 | | FKBP COMPLEXED WITH 4-HYDROXY-2-BUTANONE | | 分子名称: | 4-HYDROXY-2-BUTANONE, AMMONIUM ION, PROTEIN (FK506-BINDING PROTEIN), ... | | 著者 | Burkhard, P, Taylor, P, Walkinshaw, M.D. | | 登録日 | 1999-10-18 | | 公開日 | 1999-10-21 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | X-ray structures of small ligand-FKBP complexes provide an estimate for hydrophobic interaction energies.
J.Mol.Biol., 295, 2000
|
|
1H4V
 
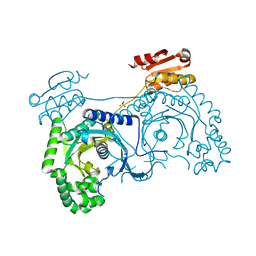 | |
228L
 
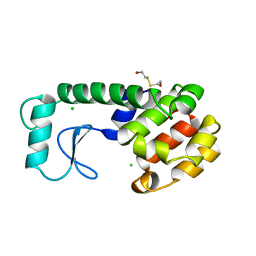 | | GENERATING LIGAND BINDING SITES IN T4 LYSOZYME USING DEFICIENCY-CREATING SUBSTITUTIONS | | 分子名称: | BETA-MERCAPTOETHANOL, CHLORIDE ION, T4 LYSOZYME | | 著者 | Baldwin, E.P, Baase, W.A, Zhang, X.-J, Feher, V, Matthews, B.W. | | 登録日 | 1997-06-25 | | 公開日 | 1998-03-18 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Generation of ligand binding sites in T4 lysozyme by deficiency-creating substitutions.
J.Mol.Biol., 277, 1998
|
|
1AKA
 
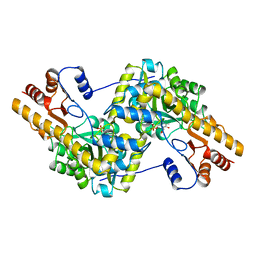 | |
1UMT
 
 | | Stromelysin-1 catalytic domain with hydrophobic inhibitor bound, ph 7.0, 32oc, 20 mm cacl2, 15% acetonitrile; nmr average of 20 structures minimized with restraints | | 分子名称: | CALCIUM ION, N-{(2R)-2-[2-(hydroxyamino)-2-oxoethyl]-4-methylpentanoyl}-L-leucyl-L-phenylalaninamide, STROMELYSIN-1, ... | | 著者 | Van Doren, S.R, Kurochkin, A.V, Hu, W, Zuiderweg, E.R.P. | | 登録日 | 1995-10-31 | | 公開日 | 1996-03-08 | | 最終更新日 | 2024-05-01 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of the catalytic domain of human stromelysin complexed with a hydrophobic inhibitor.
Protein Sci., 4, 1995
|
|
1V1A
 
 | |
1AMZ
 
 | |
1GU7
 
 | | Enoyl thioester reductase from Candida tropicalis | | 分子名称: | ENOYL-[ACYL-CARRIER-PROTEIN] REDUCTASE [NADPH, B-SPECIFIC] 1,MITOCHONDRIAL, GLYCEROL, ... | | 著者 | Airenne, T.T, Torkko, J.M, Wierenga, R.K, Hiltunen, J.K. | | 登録日 | 2002-01-24 | | 公開日 | 2003-03-13 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Structure-Function Analysis of Enoyl Thioester Reductase Involved in Mitochondrial Maintenance
J.Mol.Biol., 327, 2003
|
|
1H7B
 
 | | Structural basis for allosteric substrate specificity regulation in class III ribonucleotide reductases, native NRDD | | 分子名称: | ANAEROBIC RIBONUCLEOTIDE-TRIPHOSPHATE REDUCTASE LARGE CHAIN, PHOSPHATE ION | | 著者 | Larsson, K.-M, Andersson, J, Sjoeberg, B.-M, Nordlund, P, Logan, D.T. | | 登録日 | 2001-07-04 | | 公開日 | 2002-03-28 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.45 Å) | | 主引用文献 | Structural Basis for Allosteric Substrate Specificty Regulation in Anaerobic Ribonucleotide Reductase
Structure, 9, 2001
|
|
1YQ6
 
 | | PRD1 vertex protein P5 | | 分子名称: | Minor capsid protein | | 著者 | Merckel, M.C, Huiskonen, J.T, Goldman, A, Bamford, D.H, Tuma, R. | | 登録日 | 2005-02-01 | | 公開日 | 2005-04-26 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | The structure of the bacteriophage PRD1 spike sheds light on the evolution of viral capsid architecture.
Mol.Cell, 18, 2005
|
|
