7UTA
 
 | | CryoEM structure of Azotobacter vinelandii nitrogenase complex (2:1 FeP:MoFeP) inhibited by BeFx during catalytic N2 reduction | | 分子名称: | 3-HYDROXY-3-CARBOXY-ADIPIC ACID, ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM, ... | | 著者 | Rutledge, H.L, Cook, B.D, Nguyen, H.P.M, Tezcan, F.A, Herzik, M.A. | | 登録日 | 2022-04-26 | | 公開日 | 2022-08-17 | | 最終更新日 | 2024-02-14 | | 実験手法 | ELECTRON MICROSCOPY (2.4 Å) | | 主引用文献 | Structures of the nitrogenase complex prepared under catalytic turnover conditions.
Science, 377, 2022
|
|
7UT8
 
 | | CryoEM structure of Azotobacter vinelandii nitrogenase complex (1:1 FeP:MoFeP, ATP-bound) during catalytic N2 reduction | | 分子名称: | 3-HYDROXY-3-CARBOXY-ADIPIC ACID, ADENOSINE-5'-TRIPHOSPHATE, FE (III) ION, ... | | 著者 | Rutledge, H.L, Cook, B, Tezcan, F.A, Herzik, M.A. | | 登録日 | 2022-04-26 | | 公開日 | 2022-08-17 | | 最終更新日 | 2024-02-14 | | 実験手法 | ELECTRON MICROSCOPY (2.43 Å) | | 主引用文献 | Structures of the nitrogenase complex prepared under catalytic turnover conditions.
Science, 377, 2022
|
|
7UT9
 
 | | CryoEM structure of Azotobacter vinelandii nitrogenase complex (1:1 FeP:MoFeP, ADP/ATP-bound) during catalytic N2 reduction | | 分子名称: | 3-HYDROXY-3-CARBOXY-ADIPIC ACID, ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ... | | 著者 | Rutledge, H.L, Cook, B, Tezcan, F.A, Herzik, M.A. | | 登録日 | 2022-04-26 | | 公開日 | 2022-08-17 | | 最終更新日 | 2024-02-14 | | 実験手法 | ELECTRON MICROSCOPY (2.44 Å) | | 主引用文献 | Structures of the nitrogenase complex prepared under catalytic turnover conditions.
Science, 377, 2022
|
|
8DFC
 
 | |
8DPN
 
 | | CryoEM structure of Azotobacter vinelandii nitrogenase MoFeP during catalytic N2 reduction | | 分子名称: | 3-HYDROXY-3-CARBOXY-ADIPIC ACID, FE (III) ION, FE(8)-S(7) CLUSTER, ... | | 著者 | Rutledge, H.L, Cook, B, Tezcan, F.A, Herzik, M.A. | | 登録日 | 2022-07-15 | | 公開日 | 2022-08-17 | | 最終更新日 | 2024-02-14 | | 実験手法 | ELECTRON MICROSCOPY (2.49 Å) | | 主引用文献 | Structures of the nitrogenase complex prepared under catalytic turnover conditions.
Science, 377, 2022
|
|
1FP4
 
 | | CRYSTAL STRUCTURE OF THE ALPHA-H195Q MUTANT OF NITROGENASE | | 分子名称: | 3-HYDROXY-3-CARBOXY-ADIPIC ACID, CALCIUM ION, FE-MO-S CLUSTER, ... | | 著者 | Sorlie, M, Christiansen, J, Lemon, B.J, Peters, J.W, Dean, D.R, Hales, B.J. | | 登録日 | 2000-08-30 | | 公開日 | 2002-11-20 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Mechanistic features and structure of the nitrogenase alpha-Gln195 MoFe protein
Biochemistry, 40, 2001
|
|
8ENN
 
 | | Homocitrate-deficient nitrogenase MoFe-protein from Azotobacter vinelandii nifV knockout | | 分子名称: | CHAPSO, CITRIC ACID, FE (III) ION, ... | | 著者 | Warmack, R.A, Maggiolo, A.O, Rees, D.C. | | 登録日 | 2022-09-30 | | 公開日 | 2023-03-08 | | 最終更新日 | 2024-06-19 | | 実験手法 | ELECTRON MICROSCOPY (2.58 Å) | | 主引用文献 | Structural consequences of turnover-induced homocitrate loss in nitrogenase.
Nat Commun, 14, 2023
|
|
8ENO
 
 | | Homocitrate-deficient nitrogenase MoFe-protein from A. vinelandii nifV knockout in complex with NafT | | 分子名称: | CHAPSO, CITRIC ACID, FE (III) ION, ... | | 著者 | Warmack, R.A, Maggiolo, A.O, Rees, D.C. | | 登録日 | 2022-09-30 | | 公開日 | 2023-03-08 | | 最終更新日 | 2024-06-19 | | 実験手法 | ELECTRON MICROSCOPY (2.71 Å) | | 主引用文献 | Structural consequences of turnover-induced homocitrate loss in nitrogenase.
Nat Commun, 14, 2023
|
|
1N2C
 
 | | NITROGENASE COMPLEX FROM AZOTOBACTER VINELANDII STABILIZED BY ADP-TETRAFLUOROALUMINATE | | 分子名称: | 3-HYDROXY-3-CARBOXY-ADIPIC ACID, ADENOSINE-5'-DIPHOSPHATE, CALCIUM ION, ... | | 著者 | Schindelin, H, Kisker, C, Rees, D.C. | | 登録日 | 1997-05-02 | | 公開日 | 1997-11-12 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Structure of ADP x AIF4(-)-stabilized nitrogenase complex and its implications for signal transduction.
Nature, 387, 1997
|
|
1G21
 
 | | MGATP-BOUND AND NUCLEOTIDE-FREE STRUCTURES OF A NITROGENASE PROTEIN COMPLEX BETWEEN LEU127DEL-FE PROTEIN AND THE MOFE PROTEIN | | 分子名称: | 3-HYDROXY-3-CARBOXY-ADIPIC ACID, ADENOSINE-5'-TRIPHOSPHATE, CALCIUM ION, ... | | 著者 | Chiu, H.-J, Peters, J.W, Lanzilotta, W.N, Ryle, M.J, Seefeldt, L.C, Howard, J.B, Rees, D.C. | | 登録日 | 2000-10-16 | | 公開日 | 2001-01-31 | | 最終更新日 | 2023-08-09 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | MgATP-Bound and nucleotide-free structures of a nitrogenase protein complex between the Leu 127 Delta-Fe-protein and the MoFe-protein.
Biochemistry, 40, 2001
|
|
8BTS
 
 | |
2AFI
 
 | | Crystal Structure of MgADP bound Av2-Av1 Complex | | 分子名称: | 3-HYDROXY-3-CARBOXY-ADIPIC ACID, ADENOSINE-5'-DIPHOSPHATE, CALCIUM ION, ... | | 著者 | Tezcan, F.A, Kaiser, J.T, Mustafi, D, Walton, M.Y, Howard, J.B, Rees, D.C. | | 登録日 | 2005-07-25 | | 公開日 | 2005-09-06 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (3.1 Å) | | 主引用文献 | Nitrogenase Complexes: Multiple Docking Sites for a Nucleotide Switch Protein
Science, 309, 2005
|
|
1M1Y
 
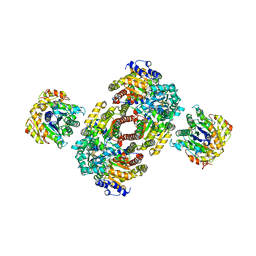 | | Chemical Crosslink of Nitrogenase MoFe Protein and Fe Protein | | 分子名称: | 3-HYDROXY-3-CARBOXY-ADIPIC ACID, CALCIUM ION, FE(8)-S(7) CLUSTER, ... | | 著者 | Schmid, B, Einsle, O, Chiu, H.J, Willing, A, Yoshida, M, Howard, J.B, Rees, D.C. | | 登録日 | 2002-06-20 | | 公開日 | 2003-02-11 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (3.2 Å) | | 主引用文献 | Biochemical and Structural Characterization of the Crosslinked Complex of Nitrogenase: Comparison to the ADP-AlF4- Stabilized Structure
Biochemistry, 41, 2002
|
|
