7Z1P
 
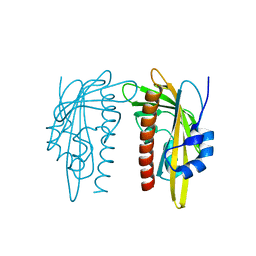 | |
7Z1S
 
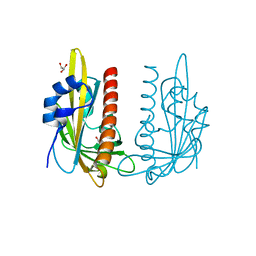 | | X-ray crystal structure of SLPYL1-NIO complex | | 分子名称: | GLYCEROL, NICOTINIC ACID, SlPYL1-NIO | | 著者 | Infantes, L, Albert, A. | | 登録日 | 2022-02-25 | | 公開日 | 2022-07-13 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | Structure-Based Modulation of the Ligand Sensitivity of a Tomato Dimeric Abscisic Acid Receptor Through a Glu to Asp Mutation in the Latch Loop.
Front Plant Sci, 13, 2022
|
|
7Z1Q
 
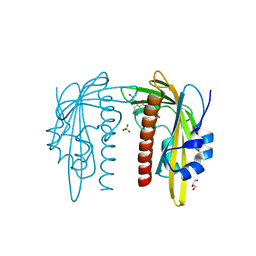 | |
7Z1R
 
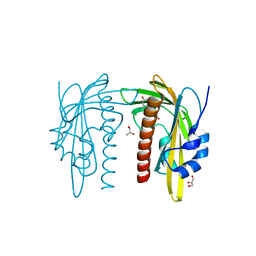 | | X-ray crystal structure of SLPYL1-E151D mutant ABA complex | | 分子名称: | (2Z,4E)-5-[(1S)-1-hydroxy-2,6,6-trimethyl-4-oxocyclohex-2-en-1-yl]-3-methylpenta-2,4-dienoic acid, GLYCEROL, SlPYL1-E151D ABA | | 著者 | Infantes, L, Albert, A. | | 登録日 | 2022-02-25 | | 公開日 | 2022-07-13 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.601 Å) | | 主引用文献 | Structure-Based Modulation of the Ligand Sensitivity of a Tomato Dimeric Abscisic Acid Receptor Through a Glu to Asp Mutation in the Latch Loop.
Front Plant Sci, 13, 2022
|
|
