1NJZ
 
 | |
1NK0
 
 | |
1NK1
 
 | | NK1 FRAGMENT OF HUMAN HEPATOCYTE GROWTH FACTOR/SCATTER FACTOR (HGF/SF) AT 2.5 ANGSTROM RESOLUTION | | 分子名称: | PROTEIN (HEPATOCYTE GROWTH FACTOR PRECURSOR) | | 著者 | Chirgadze, D.Y, Hepple, J.P, Zhou, H, Byrd, R.A, Blundell, T.L, Gherardi, E. | | 登録日 | 1998-08-20 | | 公開日 | 1999-01-13 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Crystal structure of the NK1 fragment of HGF/SF suggests a novel mode for growth factor dimerization and receptor binding.
Nat.Struct.Biol., 6, 1999
|
|
1NK2
 
 | | VND/NK-2 HOMEODOMAIN/DNA COMPLEX, NMR, 20 STRUCTURES | | 分子名称: | DNA (5'-D(*AP*CP*AP*GP*CP*CP*AP*CP*TP*TP*GP*AP*CP*AP*CP*A)-3'), DNA (5'-D(*TP*GP*TP*GP*TP*CP*AP*AP*GP*TP*GP*GP*CP*TP*GP*T)-3'), HOMEOBOX PROTEIN VND | | 著者 | Gruschus, J.M, Tsao, D.H.H, Wang, L.-H, Nirenberg, M, Ferretti, J.A. | | 登録日 | 1998-05-06 | | 公開日 | 1999-02-23 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Interactions of the vnd/NK-2 homeodomain with DNA by nuclear magnetic resonance spectroscopy: basis of binding specificity.
Biochemistry, 36, 1997
|
|
1NK3
 
 | | VND/NK-2 HOMEODOMAIN/DNA COMPLEX, NMR, MINIMIZED AVERAGE STRUCTURE | | 分子名称: | DNA (5'-D(*AP*CP*AP*GP*CP*CP*AP*CP*TP*TP*GP*AP*CP*AP*CP*A)-3'), DNA (5'-D(*TP*GP*TP*GP*TP*CP*AP*AP*GP*TP*GP*GP*CP*TP*GP*T)-3'), HOMEOBOX PROTEIN VND | | 著者 | Gruschus, J.M, Tsao, D.H.H, Wang, L.-H, Nirenberg, M, Ferretti, J.A. | | 登録日 | 1998-05-06 | | 公開日 | 1998-12-09 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Interactions of the vnd/NK-2 homeodomain with DNA by nuclear magnetic resonance spectroscopy: basis of binding specificity.
Biochemistry, 36, 1997
|
|
1NK4
 
 | |
1NK5
 
 | |
1NK6
 
 | |
1NK7
 
 | |
1NK8
 
 | | A BACILLUS DNA POLYMERASE I PRODUCT COMPLEX BOUND TO A GUANINE-THYMINE MISMATCH AFTER A SINGLE ROUND OF PRIMER EXTENSION, FOLLOWING INCORPORATION OF DCTP. | | 分子名称: | DNA POLYMERASE I, DNA PRIMER STRAND, DNA TEMPLATE STRAND, ... | | 著者 | Johnson, S.J, Beese, L.S. | | 登録日 | 2003-01-02 | | 公開日 | 2004-03-30 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Structures of mismatch replication errors observed in a DNA polymerase.
Cell(Cambridge,Mass.), 116, 2004
|
|
1NK9
 
 | | A BACILLUS DNA POLYMERASE I PRODUCT COMPLEX BOUND TO A GUANINE-THYMINE MISMATCH AFTER TWO ROUNDS OF PRIMER EXTENSION, FOLLOWING INCORPORATION OF DCTP AND DGTP. | | 分子名称: | DNA POLYMERASE I, DNA PRIMER STRAND, DNA TEMPLATE STRAND, ... | | 著者 | Johnson, S.J, Beese, L.S. | | 登録日 | 2003-01-02 | | 公開日 | 2004-03-30 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Structures of mismatch replication errors observed in a DNA polymerase.
Cell(Cambridge,Mass.), 116, 2004
|
|
1NKB
 
 | | A BACILLUS DNA POLYMERASE I PRODUCT COMPLEX BOUND TO A GUANINE-THYMINE MISMATCH AFTER THREE ROUNDS OF PRIMER EXTENSION, FOLLOWING INCORPORATION OF DCTP, DGTP, AND DTTP. | | 分子名称: | DNA POLYMERASE I, DNA PRIMER STRAND, DNA TEMPLATE STRAND, ... | | 著者 | Johnson, S.J, Beese, L.S. | | 登録日 | 2003-01-02 | | 公開日 | 2004-03-30 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structures of mismatch replication errors observed in a DNA polymerase.
Cell(Cambridge,Mass.), 116, 2004
|
|
1NKC
 
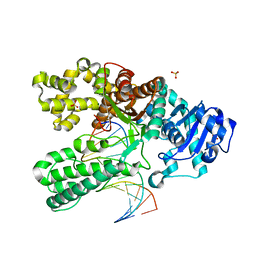 | | A BACILLUS DNA POLYMERASE I PRODUCT COMPLEX BOUND TO A GUANINE-THYMINE MISMATCH AFTER FIVE ROUNDS OF PRIMER EXTENSION, FOLLOWING INCORPORATION OF DCTP, DGTP, DTTP, AND DATP. | | 分子名称: | DNA POLYMERASE I, DNA PRIMER STRAND, DNA TEMPLATE STRAND, ... | | 著者 | Johnson, S.J, Beese, L.S. | | 登録日 | 2003-01-02 | | 公開日 | 2004-03-30 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structures of mismatch replication errors observed in a DNA polymerase.
Cell(Cambridge,Mass.), 116, 2004
|
|
1NKD
 
 | |
1NKE
 
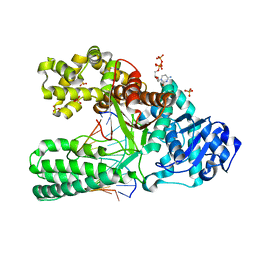 | | A BACILLUS DNA POLYMERASE I PRODUCT COMPLEX BOUND TO A CYTOSINE-THYMINE MISMATCH AFTER A SINGLE ROUND OF PRIMER EXTENSION, FOLLOWING INCORPORATION OF DCTP. | | 分子名称: | 2'-DEOXYCYTIDINE-5'-TRIPHOSPHATE, DNA POLYMERASE I, DNA PRIMER STRAND, ... | | 著者 | Johnson, S.J, Beese, L.S. | | 登録日 | 2003-01-02 | | 公開日 | 2004-03-30 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structures of mismatch replication errors observed in a DNA polymerase.
Cell(Cambridge,Mass.), 116, 2004
|
|
1NKF
 
 | |
1NKG
 
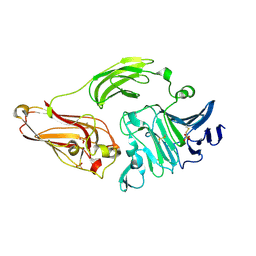 | | Rhamnogalacturonan lyase from Aspergillus aculeatus | | 分子名称: | CALCIUM ION, Rhamnogalacturonase B, SULFATE ION | | 著者 | McDonough, M.A, Kadirvelraj, R, Harris, P, Poulsen, J.C, Larsen, S. | | 登録日 | 2003-01-03 | | 公開日 | 2004-05-25 | | 最終更新日 | 2018-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Rhamnogalacturonan lyase reveals a unique three-domain modular structure for polysaccharide lyase family 4.
Febs Lett., 565, 2004
|
|
1NKH
 
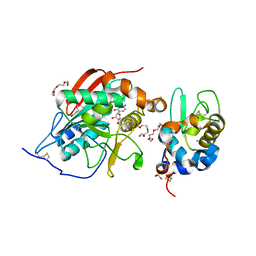 | | Crystal structure of Lactose synthase complex with UDP and Manganese | | 分子名称: | ALPHA-LACTALBUMIN, BETA-1,4-GALACTOSYLTRANSFERASE, CALCIUM ION, ... | | 著者 | Ramakrishnan, B, Qasba, P.K. | | 登録日 | 2003-01-03 | | 公開日 | 2003-01-14 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Crystal structure of lactose synthase reveals a large conformational
change in its catalytic component, the beta-1,4-galactosyltransferase
J.Mol.Biol., 310, 2001
|
|
1NKI
 
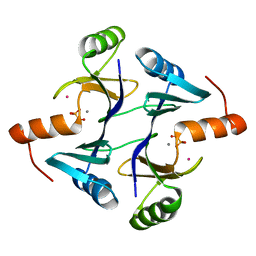 | | CRYSTAL STRUCTURE OF THE FOSFOMYCIN RESISTANCE PROTEIN A (FOSA) CONTAINING BOUND PHOSPHONOFORMATE | | 分子名称: | MANGANESE (II) ION, PHOSPHONOFORMIC ACID, POTASSIUM ION, ... | | 著者 | Rife, C.L, Pharris, R.E, Newcomer, M.E, Armstrong, R.N. | | 登録日 | 2003-01-03 | | 公開日 | 2004-01-13 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (0.95 Å) | | 主引用文献 | Phosphonoformate: a minimal transition state analogue inhibitor of the fosfomycin resistance protein, FosA.
Biochemistry, 43, 2004
|
|
1NKK
 
 | | COMPLEX STRUCTURE OF HCMV PROTEASE AND A PEPTIDOMIMETIC INHIBITOR | | 分子名称: | Capsid protein P40, Peptidomimetic inhibitor | | 著者 | Khayat, R, Batra, R, Qian, C, Halmos, T, Bailey, M, Tong, L. | | 登録日 | 2003-01-03 | | 公開日 | 2003-02-11 | | 最終更新日 | 2021-10-27 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Structural and Biochemical Studies of Inhibitor Binding to Human Cytomegalovirus Protease
Biochemistry, 42, 2003
|
|
1NKL
 
 | |
1NKM
 
 | | Complex structure of HCMV Protease and a peptidomimetic inhibitor | | 分子名称: | Assemblin, N-(6-aminohexanoyl)-3-methyl-L-valyl-3-methyl-L-valyl-N~1~-[(2S,3S)-3-hydroxy-4-oxo-4-{[(1R)-1-phenylpropyl]amino}butan-2-yl]-N~4~,N~4~-dimethyl-L-aspartamide | | 著者 | Khayat, R, Batra, R, Qian, C, Halmos, T, Bailey, M, Tong, L. | | 登録日 | 2003-01-03 | | 公開日 | 2003-02-11 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Structural and Biochemical Studies of Inhibitor Binding to Human Cytomegalovirus Protease
Biochemistry, 42, 2003
|
|
1NKN
 
 | | VISUALIZING AN UNSTABLE COILED COIL: THE CRYSTAL STRUCTURE OF AN N-TERMINAL SEGMENT OF THE SCALLOP MYOSIN ROD | | 分子名称: | S2N51-GCN4 | | 著者 | Li, Y, Brown, J.H, Reshetnikova, L, Blazsek, A, Farkas, L, Nyitray, L, Cohen, C. | | 登録日 | 2003-01-03 | | 公開日 | 2003-07-22 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Visualization of an unstable coiled coil from the scallop myosin rod
Nature, 424, 2003
|
|
1NKO
 
 | | Energetic and structural basis of sialylated oligosaccharide recognition by the natural killer cell inhibitory receptor p75/AIRM1 or Siglec-7 | | 分子名称: | Sialic acid binding Ig-like lectin 7 | | 著者 | Dimasi, N, Attril, H, van Aalten, D.M.F, Moretta, L, Biassoni, R, Mariuzza, R.A. | | 登録日 | 2003-01-03 | | 公開日 | 2003-04-01 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (1.45 Å) | | 主引用文献 | Structure of the saccharide-binding domain of the human natural killer cell inhibitory receptor p75/AIRM1.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1NKP
 
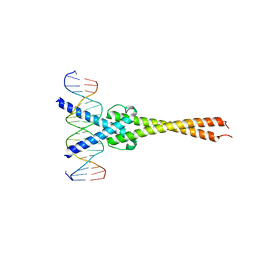 | | Crystal structure of Myc-Max recognizing DNA | | 分子名称: | 5'-D(*CP*GP*AP*GP*TP*AP*GP*CP*AP*CP*GP*TP*GP*CP*TP*AP*CP*TP*C)-3', Max protein, Myc proto-oncogene protein | | 著者 | Nair, S.K, Burley, S.K. | | 登録日 | 2003-01-03 | | 公開日 | 2003-02-04 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | X-ray structures of Myc-Max and Mad-Max recognizing DNA: Molecular bases of regulation by proto-oncogenic transcription factors
Cell(Cambridge,Mass.), 112, 2003
|
|
