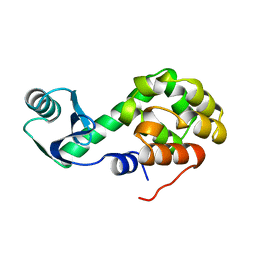
1L05
 
 | |
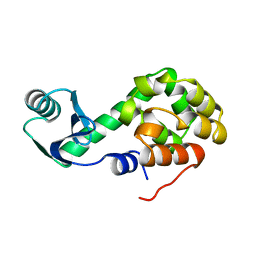
1L06
 
 | |
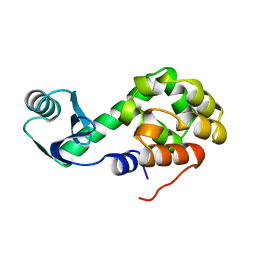
1L07
 
 | |
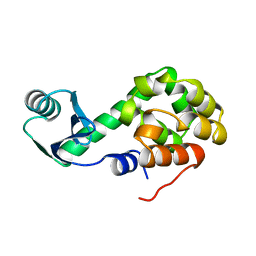
1L08
 
 | |
1L09
 
 | |
1L0A
 
 | | DOWNSTREAM REGULATOR TANK BINDS TO THE CD40 RECOGNITION SITE ON TRAF3 | | 分子名称: | TNF receptor associated factor 3, TRAF family member-associated NF-kappa-b activator | | 著者 | Li, C, Ni, C.-Z, Havert, M.L, Cabezas, E, He, J, Kaiser, D, Reed, J.C, Satterthwait, A.C, Cheng, G, Ely, K.R. | | 登録日 | 2002-02-08 | | 公開日 | 2002-04-10 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Downstream regulator TANK binds to the CD40 recognition site on TRAF3.
Structure, 10, 2002
|
|
1L0B
 
 | | Crystal Structure of rat Brca1 tandem-BRCT region | | 分子名称: | BRCA1 | | 著者 | Joo, W.S, Jeffrey, P.D, Cantor, S.B, Finnin, M.S, Livingston, D.M, Pavletich, N.P. | | 登録日 | 2002-02-08 | | 公開日 | 2002-03-20 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Structure of the 53BP1 BRCT region bound to p53 and its comparison to the Brca1 BRCT structure.
Genes Dev., 16, 2002
|
|
1L0C
 
 | | Investigation of the Roles of Catalytic Residues in Serotonin N-Acetyltransferase | | 分子名称: | COA-S-ACETYL TRYPTAMINE, Serotonin N-acetyltransferase | | 著者 | Scheibner, K.A, De Angelis, J, Burley, S.K, Cole, P.A. | | 登録日 | 2002-02-08 | | 公開日 | 2002-06-19 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Investigation of the roles of catalytic residues in serotonin N-acetyltransferase.
J.Biol.Chem., 277, 2002
|
|
1L0D
 
 | |
1L0E
 
 | |
1L0F
 
 | |
1L0G
 
 | |
1L0H
 
 | | CRYSTAL STRUCTURE OF BUTYRYL-ACP FROM E.COLI | | 分子名称: | ACYL CARRIER PROTEIN, ZINC ION | | 著者 | Roujeinikova, A, Baldock, C, Simon, W.J, Gilroy, J, Baker, P.J, Stuitje, A.R, Rice, D.W, Slabas, A.R, Rafferty, J.B. | | 登録日 | 2002-02-11 | | 公開日 | 2003-02-11 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | X-ray crystallographic studies on butyryl-ACP reveal flexibility of the structure around a putative acyl chain binding site
Structure, 10, 2002
|
|
1L0I
 
 | | Crystal structure of butyryl-ACP I62M mutant | | 分子名称: | Acyl carrier protein, CACODYLATE ION, SODIUM ION, ... | | 著者 | Roujeinikova, A, Baldock, C, Simon, W.J, Gilroy, J, Baker, P.J, Stuitje, A.R, Rice, D.W, Slabas, A.R, Rafferty, J.B. | | 登録日 | 2002-02-11 | | 公開日 | 2003-02-11 | | 最終更新日 | 2021-10-27 | | 実験手法 | X-RAY DIFFRACTION (1.2 Å) | | 主引用文献 | X-ray Crystallographic Studies on Butyryl-ACP Reveal Flexibility of the Structure around a Putative Acyl Chain Binding Site
Structure, 10, 2002
|
|
1L0J
 
 | | METHIONINE CORE MUTANT OF T4 LYSOZYME | | 分子名称: | 2-HYDROXYETHYL DISULFIDE, CHLORIDE ION, LYSOZYME | | 著者 | Gassner, N.C, Baase, W.A, Mooers, B.H, Busam, R.D, Weaver, L.H, Lindstrom, J.D, Quillin, M.L, Matthews, B.W. | | 登録日 | 2002-02-11 | | 公開日 | 2003-06-03 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.98 Å) | | 主引用文献 | Multiple methionine substitutions are tolerated in T4 lysozyme and have coupled effects on folding and stability
BIOPHYS.CHEM., 100, 2003
|
|
1L0K
 
 | | METHIONINE CORE MUTANT OF T4 LYSOZYME | | 分子名称: | 2-HYDROXYETHYL DISULFIDE, CHLORIDE ION, LYSOZYME | | 著者 | Gassner, N.C, Baase, W.A, Mooers, B.H, Busam, R.D, Weaver, L.H, Lindstrom, J.D, Quillin, M.L, Matthews, B.W. | | 登録日 | 2002-02-11 | | 公開日 | 2003-06-03 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.02 Å) | | 主引用文献 | Multiple methionine substitutions are tolerated in T4 lysozyme and have coupled effects on folding and stability
BIOPHYS.CHEM., 100, 2003
|
|
1L0L
 
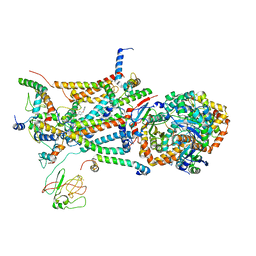 | | structure of bovine mitochondrial cytochrome bc1 complex with a bound fungicide famoxadone | | 分子名称: | Cytochrome B, Cytochrome c1, heme protein, ... | | 著者 | Gao, X, Wen, X, Yu, C.A, Esser, L, Tsao, S, Quinn, B, Zhang, L, Yu, L, Xia, D. | | 登録日 | 2002-02-11 | | 公開日 | 2003-04-08 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (2.35 Å) | | 主引用文献 | The Crystal Structure of Mitochondrial Cytochrome bc1 in Complex with Famoxadone: The Role of Aromatic-Aromatic Interaction in Inhibition
Biochemistry, 41, 2003
|
|
1L0M
 
 | |
1L0N
 
 | | native structure of bovine mitochondrial cytochrome bc1 complex | | 分子名称: | Cytochrome B, Cytochrome c1, heme protein, ... | | 著者 | Gao, X, Wen, X, Yu, C.A, Esser, L, Tsao, S, Quinn, B, Zhang, L, Yu, L, Xia, D. | | 登録日 | 2002-02-11 | | 公開日 | 2003-04-08 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | The Crystal Structure of Mitochondrial Cytochrome bc1 in Complex with Famoxadone: The Role of Aromatic-Aromatic Interaction in Inhibition
Biochemistry, 41, 2003
|
|
1L0O
 
 | | Crystal Structure of the Bacillus stearothermophilus Anti-Sigma Factor SpoIIAB with the Sporulation Sigma Factor SigmaF | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, Anti-sigma F factor, MAGNESIUM ION, ... | | 著者 | Campbell, E.A, Masuda, S, Sun, J.L, Muzzin, O, Olson, C.A, Wang, S, Darst, S.A. | | 登録日 | 2002-02-12 | | 公開日 | 2002-04-03 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Crystal structure of the Bacillus stearothermophilus anti-sigma factor SpoIIAB with the sporulation sigma factor sigmaF.
Cell(Cambridge,Mass.), 108, 2002
|
|
1L0P
 
 | |
1L0Q
 
 | | Tandem YVTN beta-propeller and PKD domains from an archaeal surface layer protein | | 分子名称: | Surface layer protein | | 著者 | Jing, H, Takagi, J, Liu, J.-H, Lindgren, S, Zhang, R.-G, Joachimiak, A, Wang, J.-H, Springer, T.A. | | 登録日 | 2002-02-12 | | 公開日 | 2002-11-06 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Archaeal Surface Layer Proteins Contain beta Propeller, PKD, and beta Helix Domains and Are Related to Metazoan Cell Surface Proteins.
Structure, 10, 2002
|
|
1L0R
 
 | |
1L0S
 
 | |
1L0V
 
 | | Quinol-Fumarate Reductase with Menaquinol Molecules | | 分子名称: | FE2/S2 (INORGANIC) CLUSTER, FE3-S4 CLUSTER, FLAVIN-ADENINE DINUCLEOTIDE, ... | | 著者 | Iverson, T.M, Luna-Chavez, C, Croal, L.R, Cecchini, G, Rees, D.C. | | 登録日 | 2002-02-13 | | 公開日 | 2002-03-13 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (3.3 Å) | | 主引用文献 | Crystallographic studies of the Escherichia coli quinol-fumarate reductase with inhibitors bound to the quinol-binding site.
J.Biol.Chem., 277, 2002
|
|
