6ZA0
 
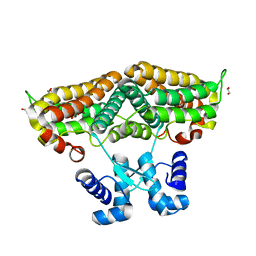 | | Structure of the transcriptional repressor Atu1419 (VanR) in complex with a fortuitous citrate from agrobacterium fabrum (P21212 space group) | | Descriptor: | 1,2-ETHANEDIOL, CITRIC ACID, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Morera, S, Vigouroux, A, Legrand, P. | | Deposit date: | 2020-06-04 | | Release date: | 2020-12-02 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Characterization of the first tetrameric transcription factor of the GntR superfamily with allosteric regulation from the bacterial pathogen Agrobacterium fabrum.
Nucleic Acids Res., 49, 2021
|
|
6ZA3
 
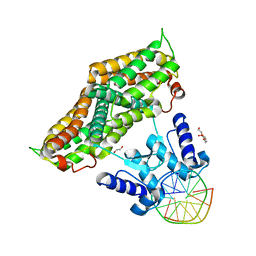 | | Structure of the transcriptional repressor Atu1419 (VanR) from agrobacterium fabrum in complex a palindromic DNA (C2221 space group) | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, CHLORIDE ION, ... | | Authors: | Morera, S, Vigouroux, A, Legrand, P. | | Deposit date: | 2020-06-04 | | Release date: | 2020-12-02 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Characterization of the first tetrameric transcription factor of the GntR superfamily with allosteric regulation from the bacterial pathogen Agrobacterium fabrum.
Nucleic Acids Res., 49, 2021
|
|
5L9P
 
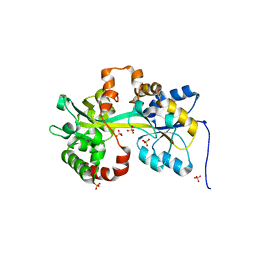 | | Crystal structure of the PBP MotA from A. tumefaciens B6 | | Descriptor: | SULFATE ION, periplasmic binding protein | | Authors: | Morera, S, Marty, L. | | Deposit date: | 2016-06-10 | | Release date: | 2016-09-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Structural Basis for High Specificity of Amadori Compound and Mannopine Opine Binding in Bacterial Pathogens.
J.Biol.Chem., 291, 2016
|
|
6R5S
 
 | |
6HQH
 
 | |
6ZK1
 
 | | Plant nucleoside hydrolase - ZmNRh2b enzyme | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Morera, S, Vigouroux, A, Kopecny, D. | | Deposit date: | 2020-06-29 | | Release date: | 2022-01-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Plant nucleoside N-ribohydrolases: riboside binding and role in nitrogen storage mobilization.
Plant J., 2023
|
|
6ZK3
 
 | | Plant nucleoside hydrolase - ZmNRh2b in complex with ribose | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Morera, S, Vigouroux, A, Kopecny, D. | | Deposit date: | 2020-06-29 | | Release date: | 2022-01-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Plant nucleoside N-ribohydrolases: riboside binding and role in nitrogen storage mobilization.
Plant J., 2023
|
|
6ZK4
 
 | | Plant nucleoside hydrolase - ZmNRh2b with a bound adenine | | Descriptor: | 1,2-ETHANEDIOL, ADENINE, CALCIUM ION, ... | | Authors: | Morera, S, Vigouroux, A, Kopecny, D. | | Deposit date: | 2020-06-29 | | Release date: | 2022-01-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Plant nucleoside N-ribohydrolases: riboside binding and role in nitrogen storage mobilization.
Plant J., 2023
|
|
6ZK5
 
 | | Plant nucleoside hydrolase - ZmNRh3 enzyme in complex with forodesine | | Descriptor: | 1,2-ETHANEDIOL, 1,4-DIDEOXY-4-AZA-1-(S)-(9-DEAZAHYPOXANTHIN-9-YL)-D-RIBITOL, CALCIUM ION, ... | | Authors: | Morera, S, Vigouroux, A, Kopecny, D. | | Deposit date: | 2020-06-29 | | Release date: | 2022-01-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Plant nucleoside N-ribohydrolases: riboside binding and role in nitrogen storage mobilization.
Plant J., 2023
|
|
6HM2
 
 | | Structure in P1 form of the PBP AgtB in complex with agropinic acid from A.tumefacien R10 | | Descriptor: | 1,2-ETHANEDIOL, Agropine permease, SODIUM ION, ... | | Authors: | Morera, S, Marty, L, Vigouroux, A. | | Deposit date: | 2018-09-12 | | Release date: | 2018-12-26 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Structural basis for two efficient modes of agropinic acid opine import into the bacterial pathogenAgrobacterium tumefaciens.
Biochem. J., 476, 2019
|
|
6HLZ
 
 | | Structure in C2 form of the PBP AgtB from A.tumefacien R10 in complex with agropinic acid | | Descriptor: | 1,2-ETHANEDIOL, 1-ETHOXY-2-(2-ETHOXYETHOXY)ETHANE, Agropine permease, ... | | Authors: | Morera, S, Marty, L, Vigouroux, A. | | Deposit date: | 2018-09-11 | | Release date: | 2018-12-26 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Structural basis for two efficient modes of agropinic acid opine import into the bacterial pathogenAgrobacterium tumefaciens.
Biochem. J., 476, 2019
|
|
6ZK2
 
 | | Plant nucleoside hydrolase - ZmNRh2b in complex with forodesine | | Descriptor: | 1,2-ETHANEDIOL, 1,4-DIDEOXY-4-AZA-1-(S)-(9-DEAZAHYPOXANTHIN-9-YL)-D-RIBITOL, CALCIUM ION, ... | | Authors: | Morera, S, Vigouroux, A, Kopecny, D. | | Deposit date: | 2020-06-29 | | Release date: | 2022-01-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Plant nucleoside N-ribohydrolases: riboside binding and role in nitrogen storage mobilization.
Plant J., 2023
|
|
6R3Z
 
 | |
6R44
 
 | |
6HLX
 
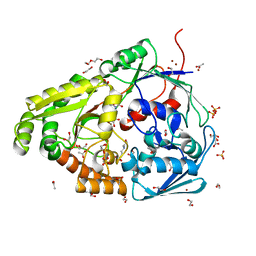 | | Structure of the PBP AgaA in complex with agropinic acid from A.tumefacien R10 | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ACETATE ION, ... | | Authors: | Morera, S, Marty, L, Vigouroux, A. | | Deposit date: | 2018-09-11 | | Release date: | 2018-12-26 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural basis for two efficient modes of agropinic acid opine import into the bacterial pathogenAgrobacterium tumefaciens.
Biochem. J., 476, 2019
|
|
1C3J
 
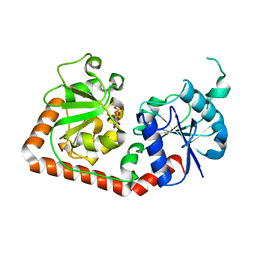 | | T4 PHAGE BETA-GLUCOSYLTRANSFERASE: SUBSTRATE BINDING AND PROPOSED CATALYTIC MECHANISM | | Descriptor: | BETA-GLUCOSYLTRANSFERASE, URIDINE-5'-DIPHOSPHATE | | Authors: | Morera, S, Imberty, A, Aschke-Sonnenborn, U, Ruger, W, Freemont, P.S. | | Deposit date: | 1999-07-28 | | Release date: | 1999-08-09 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | T4 phage beta-glucosyltransferase: substrate binding and proposed catalytic mechanism.
J.Mol.Biol., 292, 1999
|
|
4I9B
 
 | | Structure of aminoaldehyde dehydrogenase 1 from Solanum lycopersium (SlAMADH1) with a thiohemiacetal intermediate | | Descriptor: | (2-hydroxyethoxy)acetaldehyde, 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Morera, S, Vigouroux, A, Kopecny, D. | | Deposit date: | 2012-12-05 | | Release date: | 2013-02-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Plant ALDH10 family: identifying critical residues for substrate specificity and trapping a thiohemiacetal intermediate.
J.Biol.Chem., 288, 2013
|
|
4KPO
 
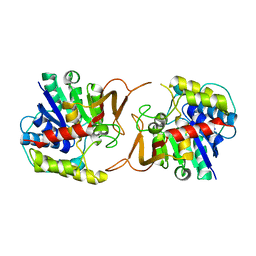 | | Plant nucleoside hydrolase - ZmNRh3 enzyme | | Descriptor: | CALCIUM ION, Nucleoside N-ribohydrolase 3 | | Authors: | Morera, S, Vigouroux, A, Kopecny, D. | | Deposit date: | 2013-05-14 | | Release date: | 2013-11-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Structure and Function of Nucleoside Hydrolases from Physcomitrella patens and Maize Catalyzing the Hydrolysis of Purine, Pyrimidine, and Cytokinin Ribosides.
Plant Physiol., 163, 2013
|
|
1QKJ
 
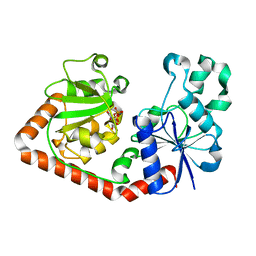 | | T4 Phage B-Glucosyltransferase, Substrate Binding and Proposed Catalytic Mechanism | | Descriptor: | BETA-GLUCOSYLTRANSFERASE, URIDINE-5'-DIPHOSPHATE | | Authors: | Morera, S, Imberty, I, Aschke-Sonnenborn, U, Ruger, W, Freemont, P.S. | | Deposit date: | 1999-07-22 | | Release date: | 1999-07-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | T4 Phage Beta-Glucosyltransferase: Substrate Binding and Proposed Catalytic Mechanism
J.Mol.Biol., 292, 1999
|
|
4I8P
 
 | | Crystal structure of aminoaldehyde dehydrogenase 1a from Zea mays (ZmAMADH1a) | | Descriptor: | 1,2-ETHANEDIOL, Aminoaldehyde dehydrogenase 1, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Morera, S, Vigouroux, A, Kopecny, D. | | Deposit date: | 2012-12-04 | | Release date: | 2013-02-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Plant ALDH10 family: identifying critical residues for substrate specificity and trapping a thiohemiacetal intermediate.
J.Biol.Chem., 288, 2013
|
|
3RX8
 
 | | structure of AaCel9A in complex with cellobiose-like isofagomine | | Descriptor: | (3R,4R,5R)-3-hydroxy-5-(hydroxymethyl)piperidin-4-yl beta-D-glucopyranoside, (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, ... | | Authors: | Morera, S. | | Deposit date: | 2011-05-10 | | Release date: | 2011-08-24 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Fortuitious binding of inhibitors-derived isofagomine for inverting GH9 beta-glycosidases
Org.Biomol.Chem., 9, 2011
|
|
4I8Q
 
 | | Structure of the aminoaldehyde dehydrogenase 1 E260A mutant from Solanum lycopersicum (SlAMADH1-E260A) | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Morera, S, Vigouroux, A, Kopecny, D. | | Deposit date: | 2012-12-04 | | Release date: | 2013-02-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Plant ALDH10 family: identifying critical residues for substrate specificity and trapping a thiohemiacetal intermediate.
J.Biol.Chem., 288, 2013
|
|
3RX5
 
 | | structure of AaCel9A in complex with cellotriose-like isofagomine | | Descriptor: | (3R,4R,5R)-3-hydroxy-5-(hydroxymethyl)piperidin-4-yl 4-O-beta-D-glucopyranosyl-beta-D-glucopyranoside, CALCIUM ION, Cellulase, ... | | Authors: | Morera, S, Vigouroux, A. | | Deposit date: | 2011-05-10 | | Release date: | 2011-08-24 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | A fortuitous binding of inhibitors-derived isofagomine for inverting GH9 beta-glycosidase
Org.Biomol.Chem., 9, 2011
|
|
3RX7
 
 | | Structure of AaCel9A in complex with cellotetraose-like isofagomine | | Descriptor: | (3R,4R,5R)-3-hydroxy-5-(hydroxymethyl)piperidin-4-yl beta-D-glucopyranoside, (3R,4R,5R)-3-hydroxy-5-(hydroxymethyl)piperidin-4-yl beta-D-glucopyranosyl-(1->4)-beta-D-glucopyranosyl-(1->4)-beta-D-glucopyranoside, CALCIUM ION, ... | | Authors: | Morera, S. | | Deposit date: | 2011-05-10 | | Release date: | 2011-08-24 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Fortuitious binding of inhibitors-derived isofagomine for inverting GH9 beta-glycosidases
Org.Biomol.Chem., 9, 2011
|
|
6RU4
 
 | |
