5ZTM
 
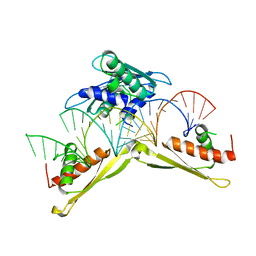 | | Crystal structure of MLE dsRBDs in complex with roX2 (R2H1) | | Descriptor: | Dosage compensation regulator, non-coding mRNA sequence roX2 | | Authors: | Lv, M.Q, Tang, Y.J. | | Deposit date: | 2018-05-04 | | Release date: | 2019-03-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.899 Å) | | Cite: | Structural insights reveal the specific recognition of roX RNA by the dsRNA-binding domains of the RNA helicase MLE and its indispensable role in dosage compensation in Drosophila.
Nucleic Acids Res., 47, 2019
|
|
8YFN
 
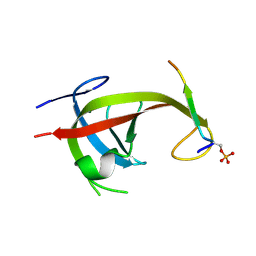 | |
8YFL
 
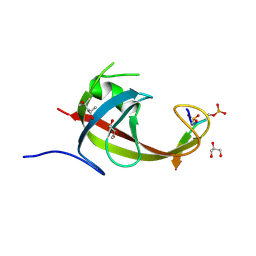 | | crystal structure of FIP200 claw/TNIP1_FIR_pS122pS123 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, GLYCEROL, RB1-inducible coiled-coil protein 1, ... | | Authors: | Lv, M.Q, Wu, S.M. | | Deposit date: | 2024-02-24 | | Release date: | 2024-08-14 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural basis for TNIP1 binding to FIP200 during mitophagy.
J.Biol.Chem., 300, 2024
|
|
8YFK
 
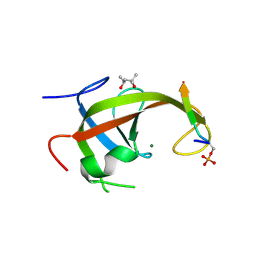 | | Crystal structure of FIP200 claw/TNIP1_FIR_pS123 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, MAGNESIUM ION, RB1-inducible coiled-coil protein 1, ... | | Authors: | Lv, M.Q, Wu, S.M. | | Deposit date: | 2024-02-24 | | Release date: | 2024-08-14 | | Last modified: | 2024-08-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for TNIP1 binding to FIP200 during mitophagy.
J.Biol.Chem., 300, 2024
|
|
8YFM
 
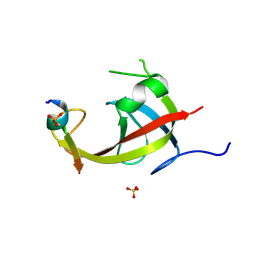 | | Crystal structure of FIP200 claw/TNIP1_FIR_pS122 | | Descriptor: | GLYCEROL, RB1-inducible coiled-coil protein 1, SULFATE ION, ... | | Authors: | Lv, M.Q, Wu, S.M. | | Deposit date: | 2024-02-24 | | Release date: | 2024-08-14 | | Last modified: | 2024-08-28 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural basis for TNIP1 binding to FIP200 during mitophagy.
J.Biol.Chem., 300, 2024
|
|
6L0X
 
 | | The First Tudor Domain of PHF20L1 | | Descriptor: | CITRIC ACID, GLYCEROL, PHD finger protein 20-like protein 1 | | Authors: | Lv, M.Q, Gao, J. | | Deposit date: | 2019-09-27 | | Release date: | 2020-09-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Conformational Selection in Ligand Recognition by the First Tudor Domain of PHF20L1.
J Phys Chem Lett, 11, 2020
|
|
6L1P
 
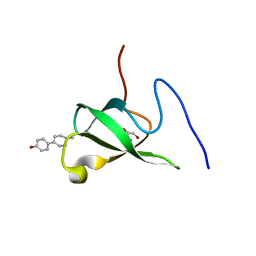 | | Crystal structure of PHF20L1 in complex with Hit 1 | | Descriptor: | 4-(1-methyl-3,6-dihydro-2H-pyridin-4-yl)phenol, GLYCEROL, PHD finger protein 20-like protein 1, ... | | Authors: | Lv, M.Q, Gao, J. | | Deposit date: | 2019-09-29 | | Release date: | 2020-09-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.231 Å) | | Cite: | Conformational Selection in Ligand Recognition by the First Tudor Domain of PHF20L1.
J Phys Chem Lett, 11, 2020
|
|
6L1C
 
 | | Crystal Structure Of of PHF20L1 Tudor1 Y24L mutant | | Descriptor: | GLYCEROL, PHD finger protein 20-like protein 1, SULFATE ION | | Authors: | Lv, M.Q, Gao, J. | | Deposit date: | 2019-09-28 | | Release date: | 2020-09-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Conformational Selection in Ligand Recognition by the First Tudor Domain of PHF20L1.
J Phys Chem Lett, 11, 2020
|
|
6L1I
 
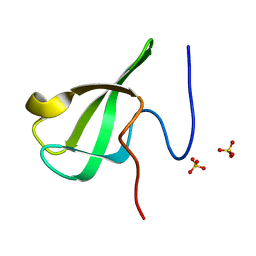 | | Crystal Structure Of of PHF20L1 Tudor1 Y24W/Y29W mutant | | Descriptor: | PHD finger protein 20-like protein 1, SULFATE ION | | Authors: | Lv, M.Q, Gao, J. | | Deposit date: | 2019-09-29 | | Release date: | 2020-09-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.849 Å) | | Cite: | Conformational Selection in Ligand Recognition by the First Tudor Domain of PHF20L1.
J Phys Chem Lett, 11, 2020
|
|
6L10
 
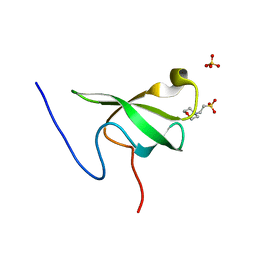 | | PHF20L1 Tudor1 - MES | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, PHD finger protein 20-like protein 1, SULFATE ION | | Authors: | Lv, M.Q, Gao, J. | | Deposit date: | 2019-09-27 | | Release date: | 2020-09-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Conformational Selection in Ligand Recognition by the First Tudor Domain of PHF20L1.
J Phys Chem Lett, 11, 2020
|
|
6L1F
 
 | |
8IPI
 
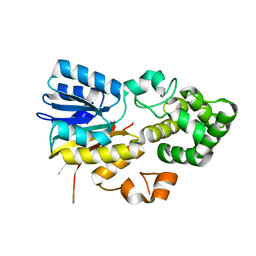 | | The apo structure of human mitochondrial methyltransferase METTL15 | | Descriptor: | 12S rRNA N4-methylcytidine (m4C) methyltransferase | | Authors: | Lv, M.Q, Zhou, W.W. | | Deposit date: | 2023-03-14 | | Release date: | 2024-01-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural insights into the specific recognition of mitochondrial ribosome-binding factor hsRBFA and 12 S rRNA by methyltransferase METTL15.
Cell Discov, 10, 2024
|
|
8IPK
 
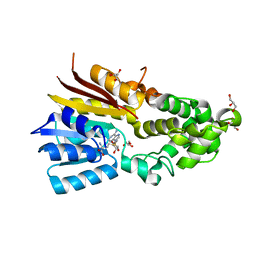 | | The structure of human mitochondrial methyltransferase METTL15 with SAM | | Descriptor: | 12S rRNA N4-methylcytidine (m4C) methyltransferase, GLYCEROL, S-ADENOSYLMETHIONINE | | Authors: | Lv, M.Q, Zhou, W.W. | | Deposit date: | 2023-03-14 | | Release date: | 2024-01-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural insights into the specific recognition of mitochondrial ribosome-binding factor hsRBFA and 12 S rRNA by methyltransferase METTL15.
Cell Discov, 10, 2024
|
|
8IPL
 
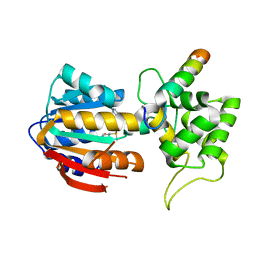 | | The structure of human mitochondrial methyltransferase METTL15 with RBFA and SAM | | Descriptor: | 12S rRNA N4-methylcytidine (m4C) methyltransferase, Putative ribosome-binding factor A, mitochondrial, ... | | Authors: | Lv, M.Q, Zhou, W.W. | | Deposit date: | 2023-03-14 | | Release date: | 2024-01-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural insights into the specific recognition of mitochondrial ribosome-binding factor hsRBFA and 12 S rRNA by methyltransferase METTL15.
Cell Discov, 10, 2024
|
|
8IPM
 
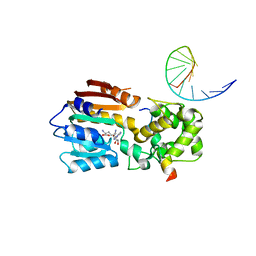 | |
8ILW
 
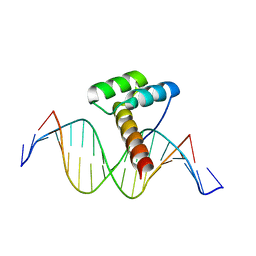 | | Transcription factor LMX1a homeobox domain in complex with Pitx3 promoter | | Descriptor: | DNA (5'-D(*CP*AP*AP*CP*AP*CP*TP*TP*AP*AP*TP*CP*CP*AP*AP*A)-3'), DNA (5'-D(*GP*TP*TP*TP*GP*GP*AP*TP*TP*AP*AP*GP*TP*GP*TP*T)-3'), LMX1A factor | | Authors: | Lv, M.Q, Lin, L.Q. | | Deposit date: | 2023-03-04 | | Release date: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.712 Å) | | Cite: | Structural insights into the recognition of the A/T-rich motif in target gene promoters by the LMX1a homeobox domain
Febs J., 2024
|
|
8IK5
 
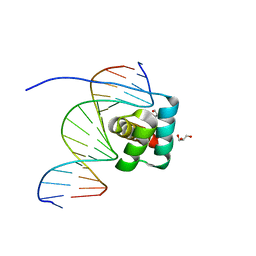 | | Transcription factor LMX1a homeobox domain in complex with Wnt1 promoter | | Descriptor: | DNA (5'-D(*CP*CP*AP*TP*AP*TP*TP*TP*AP*AP*TP*CP*TP*TP*C)-3'), DNA (5'-D(*GP*GP*AP*AP*GP*AP*TP*TP*AP*AP*AP*TP*AP*TP*G)-3'), GLYCEROL, ... | | Authors: | Lv, M.Q, Lin, L.Q. | | Deposit date: | 2023-02-28 | | Release date: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.989 Å) | | Cite: | Structural insights into the recognition of the A/T-rich motif in target gene promoters by the LMX1a homeobox domain
Febs J., 2024
|
|
8IKE
 
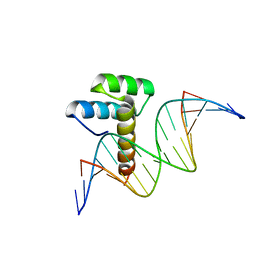 | |
7EIU
 
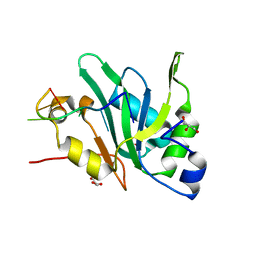 | | Crystal structure of Mei2 RRM3 in complex with 8mer meiRNA | | Descriptor: | GLYCEROL, Meiosis protein mei2, RNA (5'-R(P*UP*UP*CP*UP*GP*C)-3') | | Authors: | Shen, S.Y, Lv, M.Q. | | Deposit date: | 2021-03-31 | | Release date: | 2022-04-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.349 Å) | | Cite: | Structural insights reveal the specific recognition of meiRNA by the Mei2 protein.
J Mol Cell Biol, 14, 2022
|
|
7DTK
 
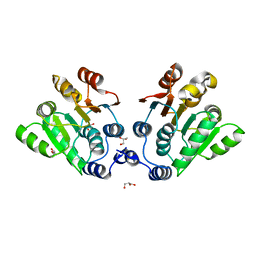 | |
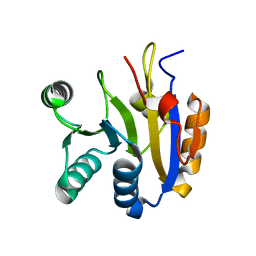
7DTJ
 
 | |
7XS4
 
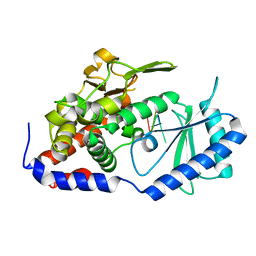 | | Crystal structure of URT1 in complex with AAAU RNA | | Descriptor: | RNA (5'-R(*AP*AP*AP*U)-3'), UTP:RNA uridylyltransferase 1 | | Authors: | Hu, Q, Zhu, L.R, lv, M.Q, Gong, Q.G. | | Deposit date: | 2022-05-12 | | Release date: | 2022-09-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.846 Å) | | Cite: | Molecular mechanism underlying the di-uridylation activity of Arabidopsis TUTase URT1.
Nucleic Acids Res., 50, 2022
|
|
