3C2E
 
 | |
3C2R
 
 | |
3C2O
 
 | |
2NLX
 
 | |
2ITM
 
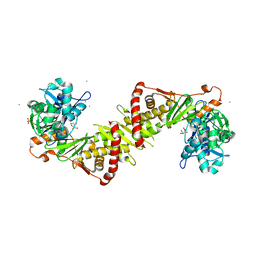 | | Crystal structure of the E. coli xylulose kinase complexed with xylulose | | Descriptor: | AMMONIUM ION, D-XYLULOSE, SULFATE ION, ... | | Authors: | di Luccio, E, Voegtli, J, Wilson, D.K. | | Deposit date: | 2006-10-19 | | Release date: | 2006-11-14 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural and kinetic studies of induced fit in xylulose kinase from Escherichia coli.
J.Mol.Biol., 365, 2007
|
|
3C2F
 
 | |
3C2V
 
 | |
4INW
 
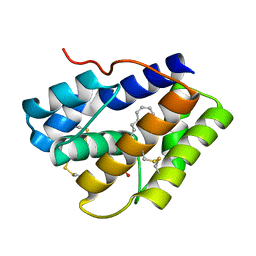 | |
4INX
 
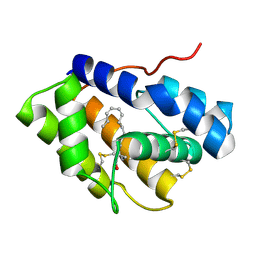 | | Structure of Pheromone-binding protein 1 in complex with (Z,Z)-11,13- hexadecadienol | | Descriptor: | (11Z,13Z)-hexadeca-11,13-dien-1-ol, Pheromone-binding protein 1 | | Authors: | di Luccio, E, Wilson, D.K. | | Deposit date: | 2013-01-07 | | Release date: | 2013-03-06 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.853 Å) | | Cite: | Crystallographic Observation of pH-Induced Conformational Changes in the Amyelois transitella Pheromone-Binding Protein AtraPBP1.
Plos One, 8, 2013
|
|
5WW0
 
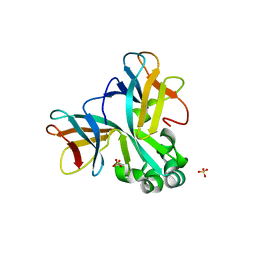 | | Crystal structure of Set7, a novel histone methyltransferase in Schizossacharomyces pombe | | Descriptor: | SET domain-containing protein 7, SULFATE ION | | Authors: | Mevius, D.E.H.F, Shen, Y, Morishita, M, Carrozzini, B, Caliandro, R, di Luccio, E. | | Deposit date: | 2016-12-30 | | Release date: | 2017-12-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Set7 Is a H3K37 Methyltransferase in Schizosaccharomyces pombe and Is Required for Proper Gametogenesis.
Structure, 27, 2019
|
|
5H6Z
 
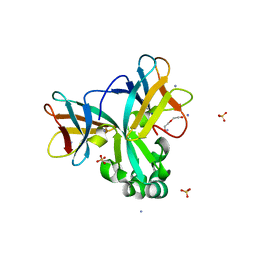 | | Crystal structure of Set7, a novel histone methyltransferase in Schizossacharomyces pombe | | Descriptor: | AMMONIUM ION, DI(HYDROXYETHYL)ETHER, SET domain-containing protein 7, ... | | Authors: | Mevius, D.E.H.F, Shen, Y, Morishita, M, Carrozzini, B, Caliandro, R, di Luccio, E. | | Deposit date: | 2016-11-15 | | Release date: | 2017-11-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Set7 Is a H3K37 Methyltransferase in Schizosaccharomyces pombe and Is Required for Proper Gametogenesis.
Structure, 27, 2019
|
|
