7NB0
 
 | | Structure of the DNA-binding domain of SEPALLATA 3 | | Descriptor: | Developmental protein SEPALLATA 3 | | Authors: | Zubieta, C, Nanao, M.H. | | Deposit date: | 2021-01-25 | | Release date: | 2021-07-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The intervening domain is required for DNA-binding and functional identity of plant MADS transcription factors.
Nat Commun, 12, 2021
|
|
8P5Q
 
 | | Structure of an ALOG domain from Arabidopsis thaliana in complex with DNA | | Descriptor: | DNA (5'-D(*AP*CP*GP*TP*CP*AP*AP*CP*AP*GP*TP*AP*AP*AP*CP*TP*A)-3'), DNA (5'-D(*TP*AP*GP*TP*TP*TP*AP*CP*TP*GP*TP*TP*GP*AP*CP*GP*T)-3'), Protein LIGHT-DEPENDENT SHORT HYPOCOTYLS 3, ... | | Authors: | Zubieta, C, Nanao, M.H, Rieu, P. | | Deposit date: | 2023-05-24 | | Release date: | 2024-02-21 | | Last modified: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | The ALOG domain defines a family of plant-specific transcription factors acting during Arabidopsis flower development.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8CRA
 
 | |
6OMS
 
 | | Arabidopsis GH3.12 with Chorismate | | Descriptor: | (3R,4R)-3-[(1-carboxyethenyl)oxy]-4-hydroxycyclohexa-1,5-diene-1-carboxylic acid, 4-substituted benzoates-glutamate ligase GH3.12, ADENOSINE MONOPHOSPHATE | | Authors: | Zubieta, C, Westfall, C.S, Holland, C.K, Jez, J.M. | | Deposit date: | 2019-04-19 | | Release date: | 2019-10-09 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.942 Å) | | Cite: | Brassicaceae-specific Gretchen Hagen 3 acyl acid amido synthetases conjugate amino acids to chorismate, a precursor of aromatic amino acids and salicylic acid.
J.Biol.Chem., 294, 2019
|
|
4OX0
 
 | |
1M6E
 
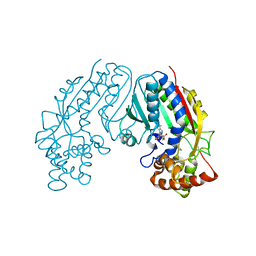 | | CRYSTAL STRUCTURE OF SALICYLIC ACID CARBOXYL METHYLTRANSFERASE (SAMT) | | Descriptor: | 2-HYDROXYBENZOIC ACID, LUTETIUM (III) ION, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Zubieta, C, Ross, J.R, Koscheski, P, Yang, Y, Pichersky, E, Noel, J.P. | | Deposit date: | 2002-07-16 | | Release date: | 2003-09-09 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural Basis for Substrate Recognition in The Salicylic Acid Carboxyl Methyltransferase Family
Plant Cell, 15, 2003
|
|
6SLR
 
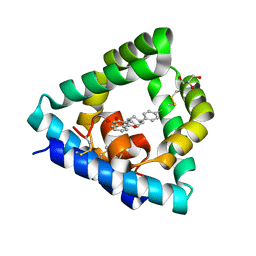 | |
2VDJ
 
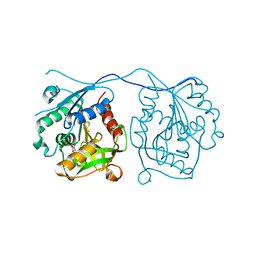 | | Crystal Structure of Homoserine O-acetyltransferase (metA) from Bacillus Cereus with Homoserine | | Descriptor: | HOMOSERINE O-SUCCINYLTRANSFERASE, L-HOMOSERINE, SULFATE ION | | Authors: | Zubieta, C, Arkus, K.A.J, Cahoon, R.E, Jez, J.M. | | Deposit date: | 2007-10-10 | | Release date: | 2008-01-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A Single Amino Acid Change is Responsible for Evolution of Acyltransferase Specificity in Bacterial Methionine Biosynthesis.
J.Biol.Chem., 283, 2008
|
|
6QEC
 
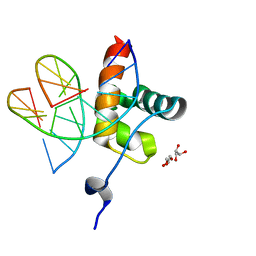 | | DNA binding domain of LUX ARRYTHMO in complex with DNA | | Descriptor: | DNA (5'-D(*AP*TP*TP*CP*GP*AP*AP*TP*AP*T*TP*AP*TP*AP*TP*TP*CP*GP*AP*A)-3'), GLYCEROL, Transcription factor LUX | | Authors: | Zubieta, C, Nayak, A. | | Deposit date: | 2019-01-07 | | Release date: | 2020-02-05 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Molecular mechanisms of Evening Complex activity inArabidopsis.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6F6O
 
 | |
4V2O
 
 | | Structure of saposin B in complex with chloroquine | | Descriptor: | N4-(7-CHLORO-QUINOLIN-4-YL)-N1,N1-DIETHYL-PENTANE-1,4-DIAMINE, SAPOSIN-B | | Authors: | Zubieta, C, Lai, X, Doyle, R.P. | | Deposit date: | 2014-10-13 | | Release date: | 2015-12-09 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | The Lysosomal Protein Saposin B Binds Chloroquine.
Chemmedchem, 11, 2016
|
|
4L39
 
 | | Crystal structure of GH3.12 from Arabidopsis thaliana in complex with AMPCPP and salicylate | | Descriptor: | 2-HYDROXYBENZOIC ACID, 4-substituted benzoates-glutamate ligase GH3.12, DIPHOSPHOMETHYLPHOSPHONIC ACID ADENOSYL ESTER, ... | | Authors: | Zubieta, C, Jez, J.M, Brown, E, Marcellin, R, Kapp, U, Round, A, Westfall, C. | | Deposit date: | 2013-06-05 | | Release date: | 2013-10-02 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Determination of the GH3.12 protein conformation through HPLC-integrated SAXS measurements combined with X-ray crystallography.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
2C6S
 
 | |
6CIG
 
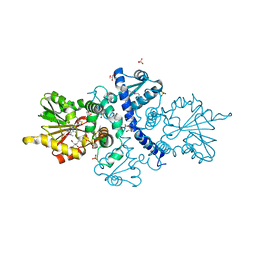 | | CRYSTAL STRUCTURE ANALYSIS OF SELENOMETHIONINE SUBSTITUTED ISOFLAVONE O-METHYLTRANSFERASE | | Descriptor: | GLYCEROL, Isoflavone-7-O-methyltransferase 8, N-(TRIS(HYDROXYMETHYL)METHYL)-3-AMINOPROPANESULFONIC ACID, ... | | Authors: | Zubieta, C, Dixon, R.A, Shabalin, I.G, Kowiel, M, Porebski, P.J, Noel, J.P. | | Deposit date: | 2018-02-23 | | Release date: | 2018-03-07 | | Last modified: | 2022-03-23 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structures of two natural product methyltransferases reveal the basis for substrate specificity in plant O-methyltransferases.
Nat. Struct. Biol., 8, 2001
|
|
1X9T
 
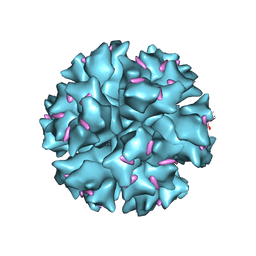 | | The crystal structure of human adenovirus 2 penton base in complex with an ad2 N-terminal fibre peptide | | Descriptor: | N-DODECYL-N,N-DIMETHYL-3-AMMONIO-1-PROPANESULFONATE, N-terminal peptide of Fiber protein, Penton protein | | Authors: | Zubieta, C, Schoehn, G, Chroboczek, J, Cusack, S. | | Deposit date: | 2004-08-24 | | Release date: | 2005-01-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | The structure of the human adenovirus 2 penton
Mol.Cell, 17, 2005
|
|
1X9P
 
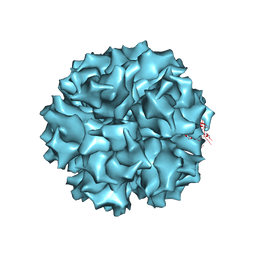 | | The crystal structure of human adenovirus 2 penton base | | Descriptor: | N-DODECYL-N,N-DIMETHYL-3-AMMONIO-1-PROPANESULFONATE, Penton protein, SULFATE ION | | Authors: | Zubieta, C, Schoehn, G, Chroboczek, J, Cusack, S. | | Deposit date: | 2004-08-24 | | Release date: | 2005-01-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | The structure of the human adenovirus 2 penton
Mol.Cell, 17, 2005
|
|
3N6Q
 
 | | Crystal structure of YghZ from E. coli | | Descriptor: | MAGNESIUM ION, YghZ aldo-keto reductase | | Authors: | Zubieta, C, Totir, M, Echols, N, May, A, Alber, T. | | Deposit date: | 2010-05-26 | | Release date: | 2011-06-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Macro-to-Micro Structural Proteomics: Native Source Proteins for High-Throughput Crystallization.
Plos One, 7, 2012
|
|
4LIY
 
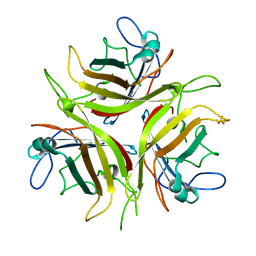 | |
1KYZ
 
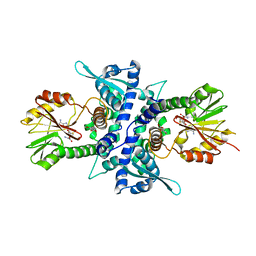 | | Crystal Structure Analysis of Caffeic acid/5-hydroxyferulic acid 3/5-O-methyltransferase Ferulic Acid Complex | | Descriptor: | 3-(4-HYDROXY-3-METHOXYPHENYL)-2-PROPENOIC ACID, Caffeic acid 3-O-methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Zubieta, C, Kota, P, Ferrer, J.-L, Dixon, R.A, Noel, J.P. | | Deposit date: | 2002-02-06 | | Release date: | 2002-08-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis for the modulation of lignin monomer methylation by caffeic acid/5-hydroxyferulic acid 3/5-O-methyltransferase.
Plant Cell, 14, 2002
|
|
1KYW
 
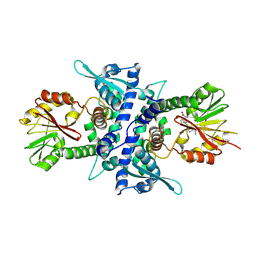 | | Crystal Structure Analysis of Caffeic Acid/5-hydroxyferulic acid 3/5-O-methyltransferase in complex with 5-hydroxyconiferaldehyde | | Descriptor: | 5-(3,3-DIHYDROXYPROPENY)-3-METHOXY-BENZENE-1,2-DIOL, Caffeic acid 3-O-methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Zubieta, C, Kota, P, Ferrer, J.-L, Dixon, R.A, Noel, J.P. | | Deposit date: | 2002-02-06 | | Release date: | 2002-08-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for the modulation of lignin monomer methylation by caffeic acid/5-hydroxyferulic acid 3/5-O-methyltransferase.
Plant Cell, 14, 2002
|
|
5LXU
 
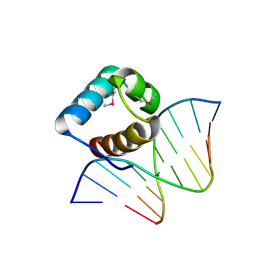 | | Structure of the DNA-binding domain of LUX ARRHYTHMO | | Descriptor: | DNA (5'-D(*AP*TP*GP*CP*GP*TP*AP*TP*CP*TP*TP*AP*GP*AP*TP*AP*CP*GP*CP*A)-3'), Transcription factor LUX | | Authors: | Zubieta, C, Nanao, M.H. | | Deposit date: | 2016-09-22 | | Release date: | 2017-10-25 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Structure of the DNA-binding domain of LUX ARRHYTHMO
To Be Published
|
|
4EQ4
 
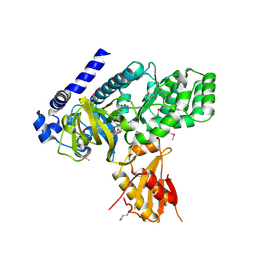 | | Crystal structure of seleno-methionine derivatized GH3.12 | | Descriptor: | 2-HYDROXYBENZOIC ACID, 4-substituted benzoates-glutamate ligase GH3.12, ADENOSINE MONOPHOSPHATE | | Authors: | Zubieta, C, Nanao, M, Jez, J, Westfall, C, Kapp, U. | | Deposit date: | 2012-04-18 | | Release date: | 2012-06-20 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.074 Å) | | Cite: | Structural basis for prereceptor modulation of plant hormones by GH3 proteins.
Science, 336, 2012
|
|
1FPQ
 
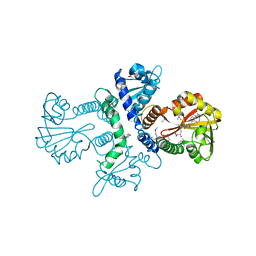 | |
1FP2
 
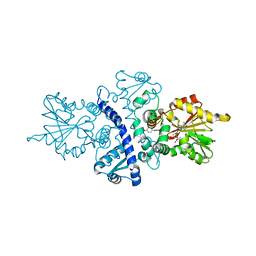 | | CRYSTAL STRUCTURE ANALYSIS OF ISOFLAVONE O-METHYLTRANSFERASE | | Descriptor: | 4'-HYDROXY-7-METHOXYISOFLAVONE, ISOFLAVONE O-METHYLTRANSFERASE, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Zubieta, C, Dixon, R.A, Noel, J.P. | | Deposit date: | 2000-08-29 | | Release date: | 2001-03-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structures of two natural product methyltransferases reveal the basis for substrate specificity in plant O-methyltransferases.
Nat.Struct.Biol., 8, 2001
|
|
1FP1
 
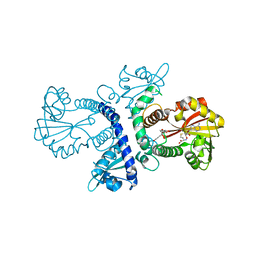 | | CRYSTAL STRUCTURE ANALYSIS OF CHALCONE O-METHYLTRANSFERASE | | Descriptor: | 2',4,4'-TRIHYDROXYCHALCONE, ISOLIQUIRITIGENIN 2'-O-METHYLTRANSFERASE, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Zubieta, C, Dixon, R.A, Noel, J.P. | | Deposit date: | 2000-08-29 | | Release date: | 2001-03-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Structures of two natural product methyltransferases reveal the basis for substrate specificity in plant O-methyltransferases.
Nat.Struct.Biol., 8, 2001
|
|
