3P3X
 
 | | Crystal Structure of the Cytochrome P450 Monooxygenase AurH (nterm-AurH-I) from Streptomyces Thioluteus | | 分子名称: | CHLORIDE ION, Cytochrome P450, GLYCEROL, ... | | 著者 | Zocher, G, Richter, M.E.A, Mueller, U, Hertweck, C. | | 登録日 | 2010-10-05 | | 公開日 | 2011-02-16 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Structural fine-tuning of a multifunctional cytochrome p450 monooxygenase.
J.Am.Chem.Soc., 133, 2011
|
|
3PH5
 
 | |
3PH6
 
 | |
7NKT
 
 | | RBD domain of SARS-CoV2 in complex with neutralizing nanobody NM1226 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, DI(HYDROXYETHYL)ETHER, PHOSPHATE ION, ... | | 著者 | Ostertag, E, Zocher, G, Stehle, T. | | 登録日 | 2021-02-18 | | 公開日 | 2021-05-12 | | 最終更新日 | 2024-10-09 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | NeutrobodyPlex-monitoring SARS-CoV-2 neutralizing immune responses using nanobodies.
Embo Rep., 22, 2021
|
|
8OQW
 
 | | Crystal structure of Tannerella forsythia MurNAc kinase MurK | | 分子名称: | ATPase, GLYCEROL, SULFATE ION | | 著者 | Gogler, K, Fink, P, Stasiak, A.C, Stehle, T, Zocher, G. | | 登録日 | 2023-04-12 | | 公開日 | 2023-08-02 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (2.05 Å) | | 主引用文献 | N-acetylmuramic acid recognition by MurK kinase from the MurNAc auxotrophic oral pathogen Tannerella forsythia.
J.Biol.Chem., 299, 2023
|
|
8OQX
 
 | | Crystal structure of Tannerella forsythia MurNAc kinase MurK with a phosphate analogue | | 分子名称: | 1,2-ETHANEDIOL, ATPase, DI(HYDROXYETHYL)ETHER, ... | | 著者 | Gogler, K, Fink, P, Stasiak, A.C, Stehle, T, Zocher, G. | | 登録日 | 2023-04-12 | | 公開日 | 2023-08-02 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (2.12 Å) | | 主引用文献 | N-acetylmuramic acid recognition by MurK kinase from the MurNAc auxotrophic oral pathogen Tannerella forsythia.
J.Biol.Chem., 299, 2023
|
|
8OW7
 
 | | Crystal structure of Tannerella forsythia sugar kinase K1058 in complex with N-acetylmuramic acid (MurNAc) | | 分子名称: | N-acetyl-beta-muramic acid, N-acetylglucosamine kinase, SULFATE ION | | 著者 | Stasiak, A.C, Gogler, K, Fink, P, Stehle, T, Zocher, G. | | 登録日 | 2023-04-27 | | 公開日 | 2023-08-02 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (3.06 Å) | | 主引用文献 | N-acetylmuramic acid recognition by MurK kinase from the MurNAc auxotrophic oral pathogen Tannerella forsythia.
J.Biol.Chem., 299, 2023
|
|
8OQK
 
 | | Crystal structure of Tannerella forsythia sugar kinase K1058 | | 分子名称: | 1,2-ETHANEDIOL, GLYCEROL, N-acetylglucosamine kinase | | 著者 | Gogler, K, Fink, P, Stasiak, A.C, Stehle, T, Zocher, G. | | 登録日 | 2023-04-12 | | 公開日 | 2023-08-02 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | N-acetylmuramic acid recognition by MurK kinase from the MurNAc auxotrophic oral pathogen Tannerella forsythia.
J.Biol.Chem., 299, 2023
|
|
8OW9
 
 | | Crystal structure of Tannerella forsythia MurNAc kinase MurK in complex with N-acetylmuramic acid (MurNAc) | | 分子名称: | N-acetyl-beta-muramic acid, Putative novel MurNAc kinase | | 著者 | Stasiak, A.C, Gogler, K, Fink, P, Stehle, T, Zocher, G. | | 登録日 | 2023-04-27 | | 公開日 | 2023-08-02 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | N-acetylmuramic acid recognition by MurK kinase from the MurNAc auxotrophic oral pathogen Tannerella forsythia.
J.Biol.Chem., 299, 2023
|
|
6GS2
 
 | | Crystal Structure of the GatD/MurT Enzyme Complex from Staphylococcus aureus | | 分子名称: | DI(HYDROXYETHYL)ETHER, MAGNESIUM ION, SA1707 protein, ... | | 著者 | Muckenfuss, L.M, Noeldeke, E.R, Niemann, V, Zocher, G, Stehle, T. | | 登録日 | 2018-06-13 | | 公開日 | 2018-09-05 | | 最終更新日 | 2024-05-15 | | 実験手法 | X-RAY DIFFRACTION (2.04 Å) | | 主引用文献 | Structural basis of cell wall peptidoglycan amidation by the GatD/MurT complex of Staphylococcus aureus.
Sci Rep, 8, 2018
|
|
3I4Z
 
 | | Crystal structure of the dimethylallyl tryptophan synthase FgaPT2 from Aspergillus fumigatus | | 分子名称: | 1,3-BUTANEDIOL, GLYCEROL, Tryptophan dimethylallyltransferase | | 著者 | Schall, C, Zocher, G, Stehle, T. | | 登録日 | 2009-07-03 | | 公開日 | 2009-09-01 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (1.76 Å) | | 主引用文献 | The structure of dimethylallyl tryptophan synthase reveals a common architecture of aromatic prenyltransferases in fungi and bacteria
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3I4X
 
 | | Crystal structure of the dimethylallyl tryptophan synthase FgaPT2 from Aspergillus fumigatus in complex with Trp and DMSPP | | 分子名称: | DIMETHYLALLYL S-THIOLODIPHOSPHATE, GLYCEROL, TRYPTOPHAN, ... | | 著者 | Schall, C, Zocher, G, Stehle, T. | | 登録日 | 2009-07-03 | | 公開日 | 2009-09-01 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | The structure of dimethylallyl tryptophan synthase reveals a common architecture of aromatic prenyltransferases in fungi and bacteria
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
4GR5
 
 | | Crystal structure of SlgN1deltaAsub in complex with AMPcPP | | 分子名称: | CHLORIDE ION, DIPHOSPHOMETHYLPHOSPHONIC ACID ADENOSYL ESTER, L(+)-TARTARIC ACID, ... | | 著者 | Herbst, D.A, Zocher, G, Stehle, T. | | 登録日 | 2012-08-24 | | 公開日 | 2012-11-28 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.92 Å) | | 主引用文献 | Structural Basis of the Interaction of MbtH-like Proteins, Putative Regulators of Nonribosomal Peptide Biosynthesis, with Adenylating Enzymes.
J.Biol.Chem., 288, 2013
|
|
4GR4
 
 | | Crystal structure of SlgN1deltaAsub | | 分子名称: | CHLORIDE ION, Non-ribosomal peptide synthetase | | 著者 | Herbst, D.A, Zocher, G, Stehle, T. | | 登録日 | 2012-08-24 | | 公開日 | 2012-11-28 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.44 Å) | | 主引用文献 | Structural Basis of the Interaction of MbtH-like Proteins, Putative Regulators of Nonribosomal Peptide Biosynthesis, with Adenylating Enzymes.
J.Biol.Chem., 288, 2013
|
|
8Q6Z
 
 | | Crystal structure of Cytochrome P450 GymB1 from Streptomyces flavidovirens | | 分子名称: | GLYCEROL, GymB1, PROTOPORPHYRIN IX CONTAINING FE | | 著者 | Freytag, J, Kelm, T, Stehle, T, Zocher, G. | | 登録日 | 2023-08-15 | | 公開日 | 2024-06-26 | | 実験手法 | X-RAY DIFFRACTION (1.96 Å) | | 主引用文献 | Intramolecular Coupling and Nucleobase Transfer - How Cytochrome P450 Enzymes GymBx Establish Their Chemoselectivity
Chemcatchem, 16, 2024
|
|
8Q6X
 
 | | Crystal structure of Cytochrome P450 GymB5 from Streptomyces katrae | | 分子名称: | Cytochrome P450, PROTOPORPHYRIN IX CONTAINING FE | | 著者 | Freytag, J, Mueller, J.M, Stehle, T, Zocher, G. | | 登録日 | 2023-08-15 | | 公開日 | 2024-06-26 | | 実験手法 | X-RAY DIFFRACTION (1.91 Å) | | 主引用文献 | Intramolecular Coupling and Nucleobase Transfer - How Cytochrome P450 Enzymes GymBx Establish Their Chemoselectivity
Chemcatchem, 16, 2024
|
|
8Q6Y
 
 | | Crystal structure of Cytochrome P450 GymB5 from Streptomyces katrae in complex with cYY and Hypoxanthine | | 分子名称: | (3S,6S)-3,6-bis(4-hydroxybenzyl)piperazine-2,5-dione, 1,2-ETHANEDIOL, Cytochrome P450, ... | | 著者 | Freytag, J, Mueller, J.M, Stehle, T, Zocher, G. | | 登録日 | 2023-08-15 | | 公開日 | 2024-06-26 | | 実験手法 | X-RAY DIFFRACTION (1.81 Å) | | 主引用文献 | Intramolecular Coupling and Nucleobase Transfer - How Cytochrome P450 Enzymes GymBx Establish Their Chemoselectivity
Chemcatchem, 16, 2024
|
|
2WBU
 
 | | CRYSTAL STRUCTURE OF THE ZINC FINGER DOMAIN OF KLF4 BOUND TO ITS TARGET DNA | | 分子名称: | 5'-D(*DGP*DAP*DGP*DGP*DCP*DGP*DTP* DGP*DGP*DC)-3', 5'-D(*DGP*DCP*DCP*DAP*DCP*DGP*DCP* DCP*DTP*DC)-3', KRUEPPEL-LIKE FACTOR 4, ... | | 著者 | Schuetz, A, Zocher, G, Carstanjen, D, Heinemann, U. | | 登録日 | 2009-03-05 | | 公開日 | 2010-04-07 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | The Structure of the Klf4 DNA-Binding Domain Links to Self-Renewal and Macrophage Differentiation.
Cell.Mol.Life Sci., 68, 2011
|
|
2WY3
 
 | | Structure of the HCMV UL16-MICB complex elucidates select binding of a viral immunoevasin to diverse NKG2D ligands | | 分子名称: | 2,5,8,11,14,17,20,23,26,29,32,35,38,41,44,47,50,53,56,59,62,65,68,71,74,77,80-HEPTACOSAOXADOOCTACONTAN-82-OL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, ... | | 著者 | Mueller, S, Zocher, G, Steinle, A, Stehle, T. | | 登録日 | 2009-11-11 | | 公開日 | 2010-02-02 | | 最終更新日 | 2024-11-06 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structure of the Hcmv Ul16-Micb Complex Elucidates Select Binding of a Viral Immunoevasin to Diverse Nkg2D Ligands.
Plos Pathog., 6, 2010
|
|
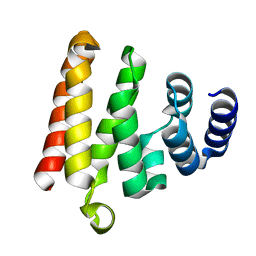
4LZV
 
 | |
4Q2S
 
 | |
4LZU
 
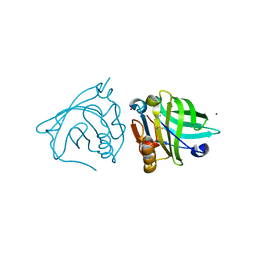 | |
4E0U
 
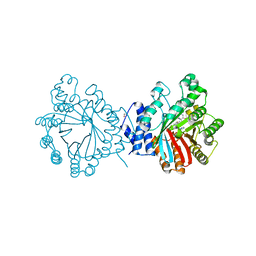 | | Crystal structure of CdpNPT in complex with thiolodiphosphate and (S)-benzodiazependione | | 分子名称: | (3S)-3-(1H-indol-3-ylmethyl)-3,4-dihydro-1H-1,4-benzodiazepine-2,5-dione, 1,2-ETHANEDIOL, Cyclic dipeptide N-prenyltransferase, ... | | 著者 | Schuller, J.M, Zocher, G, Stehle, T. | | 登録日 | 2012-03-05 | | 公開日 | 2012-05-30 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Structure and catalytic mechanism of a cyclic dipeptide prenyltransferase with broad substrate promiscuity.
J.Mol.Biol., 422, 2012
|
|
4E0T
 
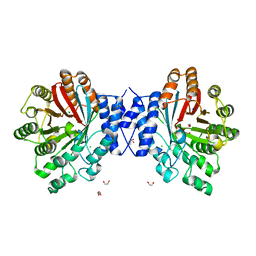 | | Crystal structure of CdpNPT in its unbound state | | 分子名称: | 1,2-ETHANEDIOL, CHLORIDE ION, Cyclic dipeptide N-prenyltransferase, ... | | 著者 | Schuller, J.M, Zocher, G, Stehle, T. | | 登録日 | 2012-03-05 | | 公開日 | 2012-05-30 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | Structure and catalytic mechanism of a cyclic dipeptide prenyltransferase with broad substrate promiscuity.
J.Mol.Biol., 422, 2012
|
|
6ZRZ
 
 | | Crystal structure of 5-dimethylallyl tryptophan synthase from Streptomyces coelicolor in complex with DMASPP and Trp | | 分子名称: | DMATS type aromatic prenyltransferase, S-(3-methylbut-2-en-1-yl) trihydrogen thiodiphosphate, TRYPTOPHAN | | 著者 | Ostertag, E, Broger, K, Stehle, T, Zocher, G. | | 登録日 | 2020-07-15 | | 公開日 | 2020-12-09 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.696 Å) | | 主引用文献 | Reprogramming Substrate and Catalytic Promiscuity of Tryptophan Prenyltransferases.
J.Mol.Biol., 433, 2020
|
|
