4M01
 
 | | N terminal fragment(residues 245-575) of binding region of SraP | | Descriptor: | CALCIUM ION, GLYCEROL, Serine-rich adhesin for platelets | | Authors: | Yang, Y.H, Jiang, Y.L, Zhang, J, Wang, L, Chen, Y, Zhou, C.Z. | | Deposit date: | 2013-08-01 | | Release date: | 2014-06-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Insights into SraP-Mediated Staphylococcus aureus Adhesion to Host Cells
Plos Pathog., 10, 2014
|
|
4E8H
 
 | | Structural of Bombyx mori glutathione transferase BmGSTD1 complex with GTT | | Descriptor: | GLUTATHIONE, Glutathione S-transferase | | Authors: | Tan, X, Ma, X.X, Hu, X.M, Chen, Q.M, Zhao, P, Xia, Q.Y, Zhou, C.Z. | | Deposit date: | 2012-03-20 | | Release date: | 2013-04-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Structural of Bombyx mori glutathione transferase BmGSTD1 complex with GTT
To be Published
|
|
4E8D
 
 | | Crystal structure of streptococcal beta-galactosidase | | Descriptor: | GLYCEROL, Glycosyl hydrolase, family 35 | | Authors: | Cheng, W, Wang, L, Bai, X.H, Jiang, Y.L, Li, Q, Yu, G, Zhou, C.Z, Chen, Y.X. | | Deposit date: | 2012-03-20 | | Release date: | 2012-05-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural insights into the substrate specificity of Streptococcus pneumoniae beta (1,3)-galactosidase BgaC
J.Biol.Chem., 287, 2012
|
|
4E8C
 
 | | Crystal structure of streptococcal beta-galactosidase in complex with galactose | | Descriptor: | GLYCEROL, Glycosyl hydrolase, family 35, ... | | Authors: | Cheng, W, Wang, L, Bai, X.H, Jiang, Y.L, Li, Q, Yu, G, Zhou, C.Z, Chen, Y.X. | | Deposit date: | 2012-03-20 | | Release date: | 2012-05-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural insights into the substrate specificity of Streptococcus pneumoniae beta (1,3)-galactosidase BgaC
J.Biol.Chem., 287, 2012
|
|
4E0H
 
 | | Crystal structure of FAD binding domain of Erv1 from Saccharomyces cerevisiae | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Mitochondrial FAD-linked sulfhydryl oxidase ERV1 | | Authors: | Guo, P.C, Ma, J.D, Jiang, Y.L, Wang, S.J, Hu, T.T, Chen, Y.X, Zhou, C.Z. | | Deposit date: | 2012-03-04 | | Release date: | 2012-08-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of yeast sulfhydryl oxidase erv1 reveals electron transfer of the disulfide relay system in the mitochondrial intermembrane space
J.Biol.Chem., 287, 2012
|
|
4E0I
 
 | | Crystal structure of the C30S/C133S mutant of Erv1 from Saccharomyces cerevisiae | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Mitochondrial FAD-linked sulfhydryl oxidase ERV1 | | Authors: | Guo, P.C, Ma, J.D, Jiang, Y.L, Wang, S.J, Hu, T.T, Chen, Y.X, Zhou, C.Z. | | Deposit date: | 2012-03-04 | | Release date: | 2012-08-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of yeast sulfhydryl oxidase erv1 reveals electron transfer of the disulfide relay system in the mitochondrial intermembrane space
J.Biol.Chem., 287, 2012
|
|
4DSS
 
 | | Crystal structure of peroxiredoxin Ahp1 from Saccharomyces cerevisiae in complex with thioredoxin Trx2 | | Descriptor: | Peroxiredoxin type-2, Thioredoxin-2 | | Authors: | Lian, F.M, Yu, J, Ma, X.X, Yu, X.J, Chen, Y, Zhou, C.Z. | | Deposit date: | 2012-02-19 | | Release date: | 2012-04-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Snapshots of Yeast Alkyl Hydroperoxide Reductase Ahp1 Peroxiredoxin Reveal a Novel Two-cysteine Mechanism of Electron Transfer to Eliminate Reactive Oxygen Species.
J.Biol.Chem., 287, 2012
|
|
4E8E
 
 | | Structural characterization of Bombyx mori glutathione transferase BmGSTD1 | | Descriptor: | Glutathione S-transferase | | Authors: | Tan, X, Ma, X.X, Hu, X.M, Chen, Q.M, Zhao, P, Xia, Q.Y, Zhou, C.Z. | | Deposit date: | 2012-03-20 | | Release date: | 2013-04-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Structural characterization of Bombyx mori glutathione transferase BmGSTD1
To be Published
|
|
4HRI
 
 | | Crystal structure of HetR in complex with a 21-bp palindromic DNA at the upstream of the hetP promoter from Anabaena | | Descriptor: | CALCIUM ION, DNA (5'-D(P*AP*TP*GP*AP*GP*GP*GP*GP*TP*TP*AP*GP*AP*CP*CP*CP*CP*TP*CP*GP*C)-3'), DNA (5'-D(P*GP*CP*GP*AP*GP*GP*GP*GP*TP*CP*TP*AP*AP*CP*CP*CP*CP*TP*CP*AP*T)-3'), ... | | Authors: | Hu, H.X, Jiang, Y.L, Zhao, M.X, Chen, Y, Zhang, C.C, Zhou, C.Z. | | Deposit date: | 2012-10-28 | | Release date: | 2013-04-17 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.954 Å) | | Cite: | Structural and biochemical analyses of Anabaena HetR reveal insights into its binding to DNA targets and the inhibitory hexapeptide ERGSGR
To be Published
|
|
4I5V
 
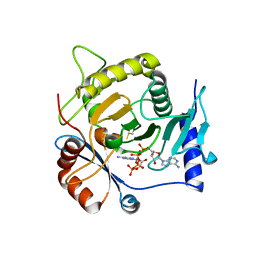 | | Crystal structure of yeast Ap4A phosphorylase Apa2 in complex with Ap4A | | Descriptor: | 5',5'''-P-1,P-4-tetraphosphate phosphorylase 2, BIS(ADENOSINE)-5'-TETRAPHOSPHATE | | Authors: | Jiang, Y.L, Hou, W.T, Chen, Y, Zhou, C.Z. | | Deposit date: | 2012-11-29 | | Release date: | 2013-05-08 | | Last modified: | 2014-02-05 | | Method: | X-RAY DIFFRACTION (2.696 Å) | | Cite: | Structures of yeast Apa2 reveal catalytic insights into a canonical AP4A phosphorylase of the histidine triad superfamily
J.Mol.Biol., 425, 2013
|
|
4I5T
 
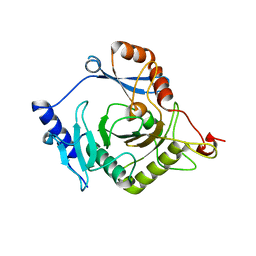 | | Crystal structure of yeast Ap4A phosphorylase Apa2 | | Descriptor: | 5',5'''-P-1,P-4-tetraphosphate phosphorylase 2 | | Authors: | Jiang, Y.L, Hou, W.T, Chen, Y, Zhou, C.Z. | | Deposit date: | 2012-11-29 | | Release date: | 2013-05-08 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structures of yeast Apa2 reveal catalytic insights into a canonical AP4A phosphorylase of the histidine triad superfamily
J.Mol.Biol., 425, 2013
|
|
4I5W
 
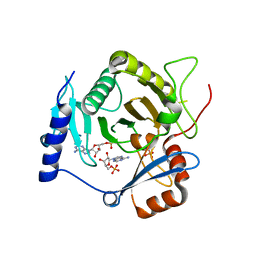 | | Crystal structure of yeast Ap4A phosphorylase Apa2 in complex with AMP | | Descriptor: | 5',5'''-P-1,P-4-tetraphosphate phosphorylase 2, ADENOSINE MONOPHOSPHATE, PHOSPHATE ION | | Authors: | Jiang, Y.L, Hou, W.T, Chen, Y, Zhou, C.Z. | | Deposit date: | 2012-11-29 | | Release date: | 2013-05-08 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.793 Å) | | Cite: | Structures of yeast Apa2 reveal catalytic insights into a canonical AP4A phosphorylase of the histidine triad superfamily
J.Mol.Biol., 425, 2013
|
|
3PPR
 
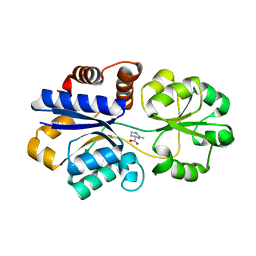 | | Structures of the substrate-binding protein provide insights into the multiple compatible solutes binding specificities of Bacillus subtilis ABC transporter OpuC | | Descriptor: | (4S)-2-METHYL-1,4,5,6-TETRAHYDROPYRIMIDINE-4-CARBOXYLIC ACID, Glycine betaine/carnitine/choline-binding protein | | Authors: | Du, Y, Shi, W.W, He, Y.X, Yang, Y.H, Zhou, C.Z, Chen, Y. | | Deposit date: | 2010-11-25 | | Release date: | 2011-05-11 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structures of the substrate-binding protein provide insights into the multiple compatible solute binding specificities of the Bacillus subtilis ABC transporter OpuC
Biochem.J., 436, 2011
|
|
3PPO
 
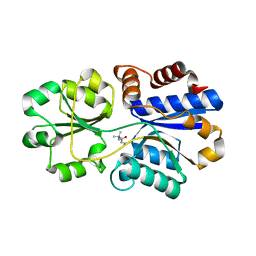 | | Structures of the substrate-binding protein provide insights into the multiple compatible solutes binding specificities of Bacillus subtilis ABC transporter OpuC | | Descriptor: | (2S)-3-carboxy-2-hydroxy-N,N,N-trimethylpropan-1-aminium, Glycine betaine/carnitine/choline-binding protein | | Authors: | Du, Y, Shi, W.W, He, Y.X, Yang, Y.H, Zhou, C.Z, Chen, Y. | | Deposit date: | 2010-11-24 | | Release date: | 2011-05-11 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structures of the substrate-binding protein provide insights into the multiple compatible solute binding specificities of the Bacillus subtilis ABC transporter OpuC
Biochem.J., 436, 2011
|
|
3PIL
 
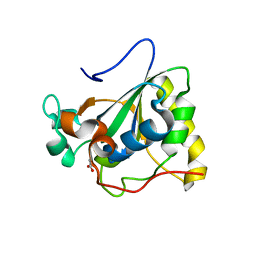 | | Crystal structure of Mxr1 from Saccharomyces cerevisiae in reduced form | | Descriptor: | ACETATE ION, Peptide methionine sulfoxide reductase | | Authors: | Ma, X.X, Guo, P.C, Shi, W.W, Luo, M, Tan, X.F, Chen, Y, Zhou, C.Z. | | Deposit date: | 2010-11-07 | | Release date: | 2011-02-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Structural plasticity of the thioredoxin recognition site of yeast methionine S-sulfoxide reductase Mxr1
J.Biol.Chem., 286, 2011
|
|
3PPP
 
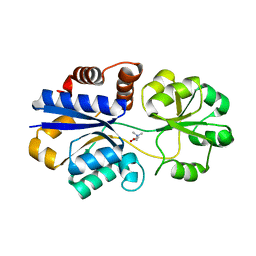 | | Structures of the substrate-binding protein provide insights into the multiple compatible solutes binding specificities of Bacillus subtilis ABC transporter OpuC | | Descriptor: | Glycine betaine/carnitine/choline-binding protein, TRIMETHYL GLYCINE | | Authors: | Du, Y, Shi, W.W, He, Y.X, Yang, Y.H, Zhou, C.Z, Chen, Y. | | Deposit date: | 2010-11-25 | | Release date: | 2011-05-11 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structures of the substrate-binding protein provide insights into the multiple compatible solute binding specificities of the Bacillus subtilis ABC transporter OpuC
Biochem.J., 436, 2011
|
|
3QWA
 
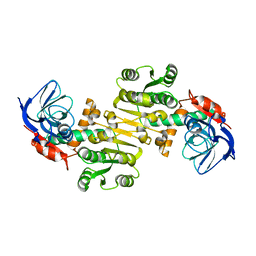 | | Crystal structure of Saccharomyces cerevisiae Zeta-crystallin-like quinone oxidoreductase Zta1 | | Descriptor: | Probable quinone oxidoreductase | | Authors: | Guo, P.C, Ma, X.X, Bao, Z.Z, Chen, Y.X, Zhou, C.Z. | | Deposit date: | 2011-02-27 | | Release date: | 2012-02-08 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural insights into the cofactor-assisted substrate recognition of yeast quinone oxidoreductase Zta1
J.Struct.Biol., 176, 2011
|
|
3QV0
 
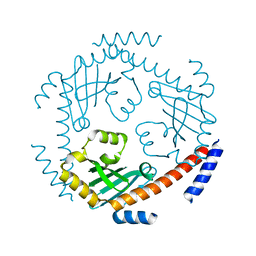 | | Crystal structure of Saccharomyces cerevisiae Mam33 | | Descriptor: | Mitochondrial acidic protein MAM33 | | Authors: | Jiang, Y.L, Pu, Y.G, Ma, X.X, Chen, Y, Zhou, C.Z. | | Deposit date: | 2011-02-24 | | Release date: | 2011-06-08 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures and putative interface of Saccharomyces cerevisiae mitochondrial matrix proteins Mmf1 and Mam33.
J.Struct.Biol., 175, 2011
|
|
3QUW
 
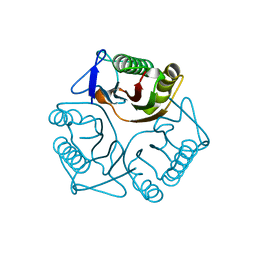 | | Crystal structure of yeast Mmf1 | | Descriptor: | Protein MMF1 | | Authors: | Jiang, Y.L, Pu, Y.G, Ma, X.X, Chen, Y, Zhou, C.Z. | | Deposit date: | 2011-02-24 | | Release date: | 2011-06-08 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structures and putative interface of Saccharomyces cerevisiae mitochondrial matrix proteins Mmf1 and Mam33.
J.Struct.Biol., 175, 2011
|
|
3QWB
 
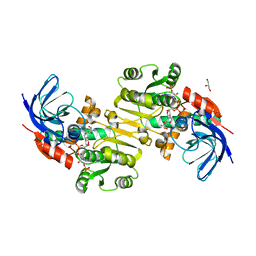 | | Crystal structure of Saccharomyces cerevisiae Zeta-crystallin-like quinone oxidoreductase Zta1 complexed with NADPH | | Descriptor: | GLYCEROL, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Probable quinone oxidoreductase | | Authors: | Guo, P.C, Ma, X.X, Bao, Z.Z, Chen, Y.X, Zhou, C.Z. | | Deposit date: | 2011-02-28 | | Release date: | 2012-02-08 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Structural insights into the cofactor-assisted substrate recognition of yeast quinone oxidoreductase Zta1
J.Struct.Biol., 176, 2011
|
|
3QFN
 
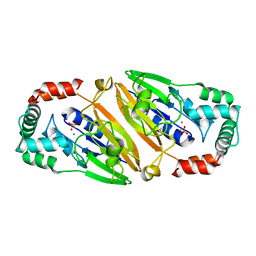 | | Crystal structure of Streptococcal asymmetric Ap4A hydrolase and phosphodiesterase Spr1479/SapH in complex with inorganic phosphate | | Descriptor: | FE (III) ION, MANGANESE (II) ION, PHOSPHATE ION, ... | | Authors: | Jiang, Y.L, Zhang, J.W, Yu, W.L, Cheng, W, Zhang, C.C, Zhou, C.Z, Chen, Y. | | Deposit date: | 2011-01-22 | | Release date: | 2011-08-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Structural and enzymatic characterization of a Streptococcal ATP/diadenosine polyphosphate and phosphodiester hydrolase Spr1479/SapH
To be Published
|
|
3PIM
 
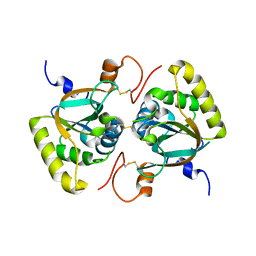 | | Crystal structure of Mxr1 from Saccharomyces cerevisiae in unusual oxidized form | | Descriptor: | Peptide methionine sulfoxide reductase | | Authors: | Ma, X.X, Guo, P.C, Shi, W.W, Luo, M, Tan, X.F, Chen, Y, Zhou, C.Z. | | Deposit date: | 2010-11-07 | | Release date: | 2011-02-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural plasticity of the thioredoxin recognition site of yeast methionine S-sulfoxide reductase Mxr1
J.Biol.Chem., 286, 2011
|
|
3PPQ
 
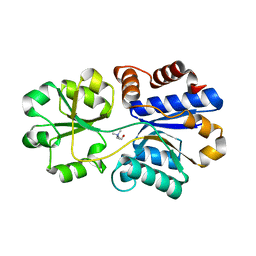 | | Structures of the substrate-binding protein provide insights into the multiple compatible solutes binding specificities of Bacillus subtilis ABC transporter OpuC | | Descriptor: | CHOLINE ION, Glycine betaine/carnitine/choline-binding protein | | Authors: | Du, Y, Shi, W.W, He, Y.X, Yang, Y.H, Zhou, C.Z, Chen, Y. | | Deposit date: | 2010-11-25 | | Release date: | 2011-05-11 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Structures of the substrate-binding protein provide insights into the multiple compatible solute binding specificities of the Bacillus subtilis ABC transporter OpuC
Biochem.J., 436, 2011
|
|
3QPM
 
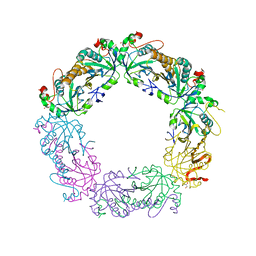 | | Crystal structure of peroxiredoxin Prx4 from Pseudosciaena crocea | | Descriptor: | GLYCEROL, Peroxiredoxin | | Authors: | Lian, F.M, Teng, Y.B, Jiang, Y.L, He, Y.X, Chen, Y, Zhou, C.Z. | | Deposit date: | 2011-02-14 | | Release date: | 2012-02-22 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The N-terminal beta-sheet of peroxiredoxin Prx4 in the large yellow croaker Pseudosciaena crocea is critical for its peroxidase and anti-bacterial activities
To be Published
|
|
3QFO
 
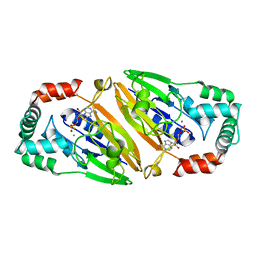 | | Crystal structure of Streptococcal asymmetric Ap4A hydrolase and phosphodiesterase Spr1479/SapH im complex with AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, FE (III) ION, MANGANESE (II) ION, ... | | Authors: | Jiang, Y.L, Zhang, J.W, Yu, W.L, Cheng, W, Zhang, C.C, Zhou, C.Z, Chen, Y. | | Deposit date: | 2011-01-22 | | Release date: | 2011-08-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and enzymatic characterization of a Streptococcal ATP/diadenosine polyphosphate and phosphodiester hydrolase Spr1479/SapH
To be Published
|
|
