8U2B
 
 | |
2MBK
 
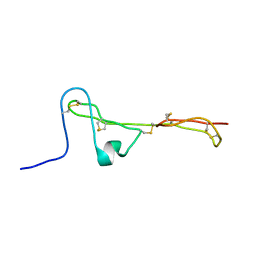 | | The Clip-segment of the von Willebrand domain 1 of the BMP modulator protein Crossveinless 2 is preformed | | Descriptor: | Crossveinless 2 | | Authors: | Mueller, T.D, Fiebig, J.E, Weidauer, S.E, Qiu, L, Bauer, M, Schmieder, P, Beerbaum, M, Zhang, J, Oschkinat, H, Sebald, W. | | Deposit date: | 2013-08-02 | | Release date: | 2013-10-16 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | The Clip-Segment of the von Willebrand Domain 1 of the BMP Modulator Protein Crossveinless 2 Is Preformed.
Molecules, 18, 2013
|
|
8HQI
 
 | |
8HQH
 
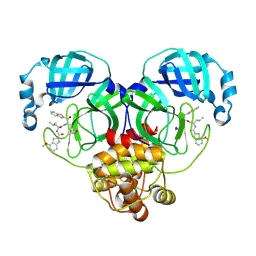 | |
8HQJ
 
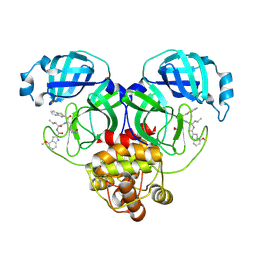 | |
8HQF
 
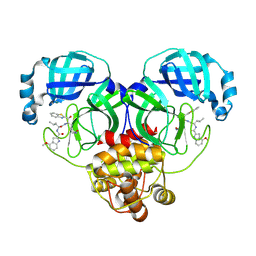 | |
8HUR
 
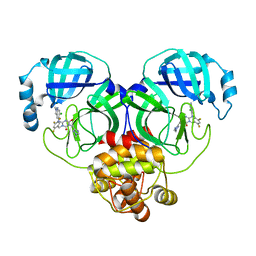 | | Crystal structure of SARS-Cov-2 main protease in complex with S217622 | | Descriptor: | 3C-like proteinase nsp5, 6-[(6-chloranyl-2-methyl-indazol-5-yl)amino]-3-[(1-methyl-1,2,4-triazol-3-yl)methyl]-1-[[2,4,5-tris(fluoranyl)phenyl]methyl]-1,3,5-triazine-2,4-dione | | Authors: | Zhou, X.L, Zhang, J, Li, J. | | Deposit date: | 2022-12-24 | | Release date: | 2023-06-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Structural basis for the inhibition of coronaviral main proteases by ensitrelvir.
Structure, 31, 2023
|
|
8HUX
 
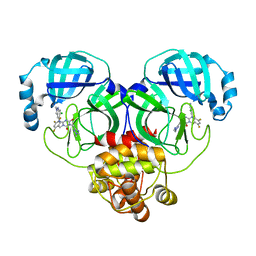 | | Crystal structure of SARS-Cov-2 main protease P132H mutant in complex with S217622 | | Descriptor: | 3C-like proteinase nsp5, 6-[(6-chloranyl-2-methyl-indazol-5-yl)amino]-3-[(1-methyl-1,2,4-triazol-3-yl)methyl]-1-[[2,4,5-tris(fluoranyl)phenyl]methyl]-1,3,5-triazine-2,4-dione | | Authors: | Li, W.W, Zhang, J, Li, J. | | Deposit date: | 2022-12-24 | | Release date: | 2023-06-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Structural basis for the inhibition of coronaviral main proteases by ensitrelvir.
Structure, 31, 2023
|
|
8HUS
 
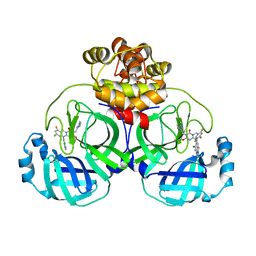 | | Crystal structure of SARS main protease in complex with S217622 | | Descriptor: | 3C-like proteinase nsp5, 6-[(6-chloranyl-2-methyl-indazol-5-yl)amino]-3-[(1-methyl-1,2,4-triazol-3-yl)methyl]-1-[[2,4,5-tris(fluoranyl)phenyl]methyl]-1,3,5-triazine-2,4-dione | | Authors: | Lin, C, Zhang, J, Li, J. | | Deposit date: | 2022-12-24 | | Release date: | 2023-06-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structural basis for the inhibition of coronaviral main proteases by ensitrelvir.
Structure, 31, 2023
|
|
8HUT
 
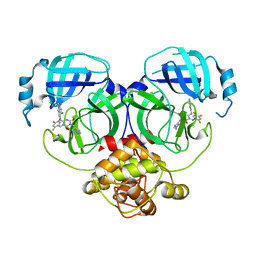 | | Crystal structure of MERS main protease in complex with S217622 | | Descriptor: | 6-[(6-chloranyl-2-methyl-indazol-5-yl)amino]-3-[(1-methyl-1,2,4-triazol-3-yl)methyl]-1-[[2,4,5-tris(fluoranyl)phenyl]methyl]-1,3,5-triazine-2,4-dione, ORF1a | | Authors: | Lin, C, Zhang, J, Li, J. | | Deposit date: | 2022-12-24 | | Release date: | 2023-06-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structural basis for the inhibition of coronaviral main proteases by ensitrelvir.
Structure, 31, 2023
|
|
8HUV
 
 | | Crystal structure of SARS-Cov-2 main protease G15S mutant in complex with S217622 | | Descriptor: | 3C-like proteinase nsp5, 6-[(6-chloranyl-2-methyl-indazol-5-yl)amino]-3-[(1-methyl-1,2,4-triazol-3-yl)methyl]-1-[[2,4,5-tris(fluoranyl)phenyl]methyl]-1,3,5-triazine-2,4-dione | | Authors: | Zeng, P, Zhang, J, Li, J. | | Deposit date: | 2022-12-24 | | Release date: | 2023-06-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structural basis for the inhibition of coronaviral main proteases by ensitrelvir.
Structure, 31, 2023
|
|
8HUU
 
 | | Crystal structure of HCoV-NL63 main protease with S217622 | | Descriptor: | 3C-like proteinase, 6-[(6-chloranyl-2-methyl-indazol-5-yl)amino]-3-[(1-methyl-1,2,4-triazol-3-yl)methyl]-1-[[2,4,5-tris(fluoranyl)phenyl]methyl]-1,3,5-triazine-2,4-dione | | Authors: | Zeng, X.Y, Zhang, J, Li, J. | | Deposit date: | 2022-12-24 | | Release date: | 2023-06-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Structural basis for the inhibition of coronaviral main proteases by ensitrelvir.
Structure, 31, 2023
|
|
8HVL
 
 | | Crystal structure of SARS-Cov-2 main protease M49I mutant in complex with PF07321332 | | Descriptor: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5 | | Authors: | Zou, X.F, Zhang, J, Li, J. | | Deposit date: | 2022-12-27 | | Release date: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Crystal structure of SARS-Cov-2 main protease
M49I mutant in complex with PF07321332
To Be Published
|
|
8HVO
 
 | | Crystal structure of SARS-Cov-2 main protease V186F mutant in complex with PF07321332 | | Descriptor: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5 | | Authors: | Zou, X.F, Zhang, J, Li, J. | | Deposit date: | 2022-12-27 | | Release date: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of SARS-Cov-2 main protease
V186F mutant in complex with PF07321332
To Be Published
|
|
8HVK
 
 | | Crystal structure of SARS-Cov-2 main protease G15S mutant in complex with PF07321332 | | Descriptor: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5 | | Authors: | Zeng, P, Zhang, J, Li, J. | | Deposit date: | 2022-12-27 | | Release date: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Crystal structure of SARS-Cov-2 main protease
G15S mutant in complex with PF07321332
To Be Published
|
|
8HVN
 
 | | Crystal structure of SARS-Cov-2 main protease P132H mutant in complex with PF07321332 | | Descriptor: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5 | | Authors: | Li, W.W, Zhang, J, Li, J. | | Deposit date: | 2022-12-27 | | Release date: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of SARS-Cov-2 main protease
P132H mutant in complex with PF07321332
To Be Published
|
|
8HZR
 
 | | Crystal structure of SARS-Cov-2 main protease S46F mutant in complex with PF07321332 | | Descriptor: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5 | | Authors: | Zeng, X.Y, Zhang, J, Li, J. | | Deposit date: | 2023-01-09 | | Release date: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Crystal structure of SARS-Cov-2 main protease
S46F mutant in complex with PF07321332
To Be Published
|
|
8UEJ
 
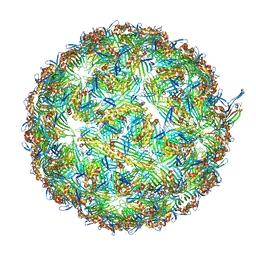 | | ssRNA phage PhiCb5 virion | | Descriptor: | CALCIUM ION, Coat protein, Maturation protein | | Authors: | Wang, Y, Zhang, J. | | Deposit date: | 2023-10-01 | | Release date: | 2024-05-15 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structural mechanisms of Tad pilus assembly and its interaction with an RNA virus.
Sci Adv, 10, 2024
|
|
8UCR
 
 | |
3V7E
 
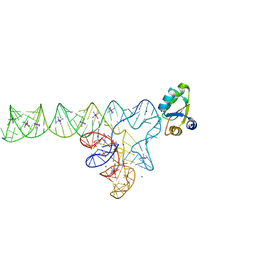 | | Crystal structure of YbxF bound to the SAM-I riboswitch aptamer | | Descriptor: | COBALT HEXAMMINE(III), MAGNESIUM ION, Ribosome-associated protein L7Ae-like, ... | | Authors: | Baird, N.J, Zhang, J, Hamma, T, Ferre-D'Amare, A.R. | | Deposit date: | 2011-12-21 | | Release date: | 2012-03-07 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | YbxF and YlxQ are bacterial homologs of L7Ae and bind K-turns but not K-loops.
Rna, 18, 2012
|
|
3VHZ
 
 | |
7DYS
 
 | |
3VI0
 
 | |
3V7Q
 
 | | Crystal structure of B. subtilis YlxQ at 1.55 A resolution | | Descriptor: | CITRIC ACID, POTASSIUM ION, Probable ribosomal protein ylxQ | | Authors: | Baird, N.J, Zhang, J, Hamma, T, Ferre-D'Amare, A.R. | | Deposit date: | 2011-12-21 | | Release date: | 2012-03-07 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | YbxF and YlxQ are bacterial homologs of L7Ae and bind K-turns but not K-loops.
Rna, 18, 2012
|
|
3ZDA
 
 | | Structure of E. coli ExoIX in complex with a fragment of the Flap1 DNA oligonucleotide, potassium and magnesium | | Descriptor: | 5'-D(*AP*AP*GP*CP*GP*CP)-3', 5'-D(*GP*CP*GP*CP)-3', MAGNESIUM ION, ... | | Authors: | Hemsworth, G.R, Anstey-Gilbert, C.S, Flemming, C.S, Hodskinson, M.R.G, Zhang, J, Sedelnikova, S.E, Stillman, T.J, Sayers, J.R, Artymiuk, P.J. | | Deposit date: | 2012-11-26 | | Release date: | 2013-07-10 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The Structure of E. Coli Exoix - Implications for DNA Binding and Catalysis in Flap Endonucleases
Nucleic Acids Res., 41, 2013
|
|
