6PTO
 
 | | Structure of Ctf4 trimer in complex with three CMG helicases | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, Cell division control protein 45, DNA polymerase alpha-binding protein, ... | | 著者 | Yuan, Z, Georgescu, R, Bai, L, Santos, R, Donnell, M, Li, H. | | 登録日 | 2019-07-16 | | 公開日 | 2019-11-20 | | 最終更新日 | 2019-12-18 | | 実験手法 | ELECTRON MICROSCOPY (7 Å) | | 主引用文献 | Ctf4 organizes sister replisomes and Pol alpha into a replication factory.
Elife, 8, 2019
|
|
6PTN
 
 | | Structure of Ctf4 trimer in complex with two CMG helicases | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, Cell division control protein 45, DNA polymerase alpha-binding protein, ... | | 著者 | Yuan, Z, Georgescu, R, Bai, L, Santos, R, Donnell, M, Li, H. | | 登録日 | 2019-07-16 | | 公開日 | 2019-11-20 | | 最終更新日 | 2019-12-18 | | 実験手法 | ELECTRON MICROSCOPY (5.8 Å) | | 主引用文献 | Ctf4 organizes sister replisomes and Pol alpha into a replication factory.
Elife, 8, 2019
|
|
6U0M
 
 | | Structure of the S. cerevisiae replicative helicase CMG in complex with a forked DNA | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, Cell division control protein 45, DNA (15-MER), ... | | 著者 | Yuan, Z, Georgescu, R, Bai, L, Zhang, D, O'Donnell, M, Li, H. | | 登録日 | 2019-08-14 | | 公開日 | 2020-03-25 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (3.9 Å) | | 主引用文献 | DNA unwinding mechanism of a eukaryotic replicative CMG helicase.
Nat Commun, 11, 2020
|
|
6PTJ
 
 | | Structure of Ctf4 trimer in complex with one CMG helicase | | 分子名称: | Cell division control protein 45, DNA polymerase alpha-binding protein, DNA replication complex GINS protein PSF1, ... | | 著者 | Yuan, Z, Georgescu, R, Bai, L, Santos, R, Donnell, M, Li, H. | | 登録日 | 2019-07-15 | | 公開日 | 2019-11-20 | | 最終更新日 | 2019-12-18 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | Ctf4 organizes sister replisomes and Pol alpha into a replication factory.
Elife, 8, 2019
|
|
4FZ4
 
 | | Crystal structure of HP0197-18kd | | 分子名称: | CHLORIDE ION, NITRATE ION, Uncharacterized protein conserved in bacteria | | 著者 | Yuan, Z, Yan, X. | | 登録日 | 2012-07-06 | | 公開日 | 2012-12-05 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.44 Å) | | 主引用文献 | Molecular mechanism by which surface antigen HP0197 mediates host cell attachment in the pathogenic bacteria Streptococcus suis
J.Biol.Chem., 288, 2013
|
|
4FZQ
 
 | | Crystal structure of HP0197-G5 | | 分子名称: | Uncharacterized protein conserved in bacteria | | 著者 | Yuan, Z, Yan, X. | | 登録日 | 2012-07-07 | | 公開日 | 2012-12-05 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Molecular mechanism by which surface antigen HP0197 mediates host cell attachment in the pathogenic bacteria Streptococcus suis
J.Biol.Chem., 288, 2013
|
|
6WGG
 
 | | Atomic model of pre-insertion mutant OCCM-DNA complex(ORC-Cdc6-Cdt1-Mcm2-7 with Mcm6 WHD truncation) | | 分子名称: | Cell division control protein 6, Cell division cycle protein CDT1, DNA (41-MER), ... | | 著者 | Yuan, Z, Schneider, S, Dodd, T, Riera, A, Bai, L, Yan, C, Magdalou, I, Ivanov, I, Stillman, B, Li, H, Speck, C. | | 登録日 | 2020-04-05 | | 公開日 | 2020-07-15 | | 最終更新日 | 2024-03-06 | | 実験手法 | ELECTRON MICROSCOPY (8.1 Å) | | 主引用文献 | Structural mechanism of helicase loading onto replication origin DNA by ORC-Cdc6.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6WGC
 
 | | Atomic model of semi-attached mutant OCCM-DNA complex (ORC-Cdc6-Cdt1-Mcm2-7 with Mcm6 WHD truncation) | | 分子名称: | Cell division control protein 6, DNA (41-MER), DNA replication licensing factor MCM3, ... | | 著者 | Yuan, Z, Schneider, S, Dodd, T, Riera, A, Bai, L, Yan, C, Magdalou, I, Ivanov, I, Stillman, B, Li, H, Speck, C. | | 登録日 | 2020-04-05 | | 公開日 | 2020-07-15 | | 最終更新日 | 2024-03-06 | | 実験手法 | ELECTRON MICROSCOPY (4.3 Å) | | 主引用文献 | Structural mechanism of helicase loading onto replication origin DNA by ORC-Cdc6.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6WGF
 
 | | Atomic model of mutant Mcm2-7 hexamer with Mcm6 WHD truncation | | 分子名称: | DNA replication licensing factor MCM2, DNA replication licensing factor MCM3, DNA replication licensing factor MCM4, ... | | 著者 | Yuan, Z, Schneider, S, Dodd, T, Riera, A, Bai, L, Yan, C, Magdalou, I, Ivanov, I, Stillman, B, Li, H, Speck, C. | | 登録日 | 2020-04-05 | | 公開日 | 2020-07-15 | | 最終更新日 | 2024-03-06 | | 実験手法 | ELECTRON MICROSCOPY (7.7 Å) | | 主引用文献 | Structural mechanism of helicase loading onto replication origin DNA by ORC-Cdc6.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6WGI
 
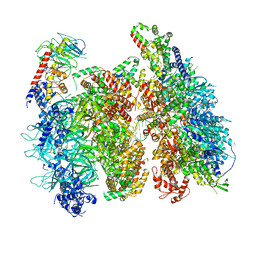 | | Atomic model of the mutant OCCM (ORC-Cdc6-Cdt1-Mcm2-7 with Mcm6 WHD truncation) loaded on DNA at 10.5 A resolution | | 分子名称: | Cell division control protein 6, Cell division cycle protein CDT1, DNA (34-MER), ... | | 著者 | Yuan, Z, Schneider, S, Dodd, T, Riera, A, Bai, L, Yan, C, Magdalou, I, Ivanov, I, Stillman, B, Li, H, Speck, C. | | 登録日 | 2020-04-05 | | 公開日 | 2020-07-15 | | 最終更新日 | 2024-03-06 | | 実験手法 | ELECTRON MICROSCOPY (10 Å) | | 主引用文献 | Structural mechanism of helicase loading onto replication origin DNA by ORC-Cdc6.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6WJV
 
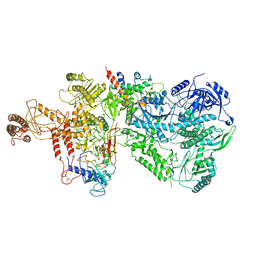 | | Structure of the Saccharomyces cerevisiae polymerase epsilon holoenzyme | | 分子名称: | DNA polymerase epsilon catalytic subunit A, DNA polymerase epsilon subunit B, DNA polymerase epsilon subunit C, ... | | 著者 | Yuan, Z, Georgescu, R, Schauer, G.D, O'Donnell, M, Li, H. | | 登録日 | 2020-04-14 | | 公開日 | 2020-07-08 | | 実験手法 | ELECTRON MICROSCOPY (3.5 Å) | | 主引用文献 | Structure of the polymerase epsilon holoenzyme and atomic model of the leading strand replisome.
Nat Commun, 11, 2020
|
|
5V8F
 
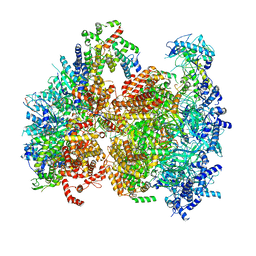 | | Structural basis of MCM2-7 replicative helicase loading by ORC-Cdc6 and Cdt1 | | 分子名称: | Cell division control protein 6, Cell division cycle protein CDT1, DNA (39-MER), ... | | 著者 | Yuan, Z, Riera, A, Bai, L, Sun, J, Spanos, C, Chen, Z.A, Barbon, M, Rappsilber, J, Stillman, B, Speck, C, Li, H. | | 登録日 | 2017-03-21 | | 公開日 | 2017-05-10 | | 最終更新日 | 2020-04-22 | | 実験手法 | ELECTRON MICROSCOPY (3.9 Å) | | 主引用文献 | Structural basis of Mcm2-7 replicative helicase loading by ORC-Cdc6 and Cdt1.
Nat. Struct. Mol. Biol., 24, 2017
|
|
8FOJ
 
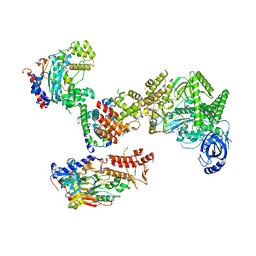 | | Cryo-EM structure of S. cerevisiae DNA polymerase alpha-primase complex in the post RNA handoff state | | 分子名称: | DNA polymerase, DNA polymerase alpha subunit B, DNA primase, ... | | 著者 | Yuan, Z, Georgescu, R, Li, H, O'Donnell, M. | | 登録日 | 2022-12-30 | | 公開日 | 2023-05-31 | | 最終更新日 | 2024-03-13 | | 実験手法 | ELECTRON MICROSCOPY (4.8 Å) | | 主引用文献 | Molecular choreography of primer synthesis by the eukaryotic Pol alpha-primase.
Nat Commun, 14, 2023
|
|
8FOH
 
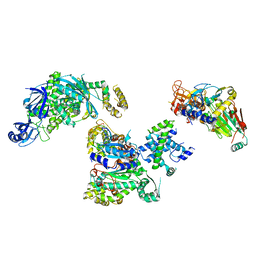 | | Cryo-EM structure of S. cerevisiae DNA polymerase alpha-primase complex in the RNA synthesis state | | 分子名称: | DNA polymerase, DNA polymerase alpha subunit B, DNA primase, ... | | 著者 | Yuan, Z, Georgescu, R, Li, H, O'Donnell, M. | | 登録日 | 2022-12-30 | | 公開日 | 2023-05-31 | | 最終更新日 | 2024-03-13 | | 実験手法 | ELECTRON MICROSCOPY (4.6 Å) | | 主引用文献 | Molecular choreography of primer synthesis by the eukaryotic Pol alpha-primase.
Nat Commun, 14, 2023
|
|
8FOC
 
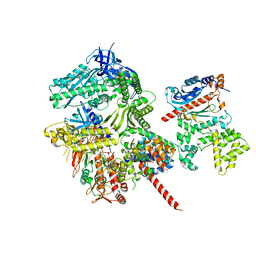 | | Cryo-EM structure of S. cerevisiae DNA polymerase alpha-primase in Apo state conformation I | | 分子名称: | DNA polymerase, DNA polymerase alpha subunit B, DNA primase, ... | | 著者 | Yuan, Z, Georgescu, R, Li, H, O'Donnell, M. | | 登録日 | 2022-12-30 | | 公開日 | 2023-05-31 | | 最終更新日 | 2024-03-13 | | 実験手法 | ELECTRON MICROSCOPY (3.7 Å) | | 主引用文献 | Molecular choreography of primer synthesis by the eukaryotic Pol alpha-primase.
Nat Commun, 14, 2023
|
|
8FOE
 
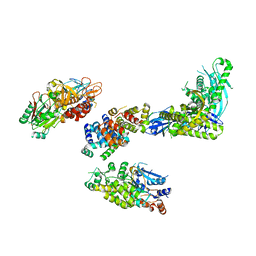 | | Cryo-EM structure of S. cerevisiae DNA polymerase alpha-primase complex bound to a template DNA | | 分子名称: | DNA polymerase, DNA polymerase alpha subunit B, DNA primase, ... | | 著者 | Yuan, Z, Georgescu, R, Li, H, O'Donnell, M. | | 登録日 | 2022-12-30 | | 公開日 | 2023-05-31 | | 最終更新日 | 2024-03-13 | | 実験手法 | ELECTRON MICROSCOPY (5.6 Å) | | 主引用文献 | Molecular choreography of primer synthesis by the eukaryotic Pol alpha-primase.
Nat Commun, 14, 2023
|
|
8FOK
 
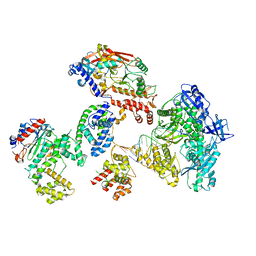 | | Cryo-EM structure of S. cerevisiae DNA polymerase alpha-primase complex in the DNA elongation state | | 分子名称: | DNA polymerase, DNA polymerase alpha subunit B, DNA primase, ... | | 著者 | Yuan, Z, Georgescu, R, Li, H, O'Donnell, M. | | 登録日 | 2022-12-30 | | 公開日 | 2023-05-31 | | 最終更新日 | 2024-03-13 | | 実験手法 | ELECTRON MICROSCOPY (3.56 Å) | | 主引用文献 | Molecular choreography of primer synthesis by the eukaryotic Pol alpha-primase.
Nat Commun, 14, 2023
|
|
8FOD
 
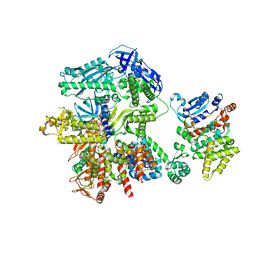 | | Cryo-EM structure of S. cerevisiae DNA polymerase alpha-primase complex in Apo state conformation II | | 分子名称: | DNA polymerase, DNA polymerase alpha subunit B, DNA primase, ... | | 著者 | Yuan, Z, Georgescu, R, Li, H, O'Donnell, M. | | 登録日 | 2022-12-30 | | 公開日 | 2023-05-31 | | 最終更新日 | 2024-03-13 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | Molecular choreography of primer synthesis by the eukaryotic Pol alpha-primase.
Nat Commun, 14, 2023
|
|
6IYF
 
 | | Structure of pSTING complex | | 分子名称: | (2R,3R,3aS,5R,7aR,9R,10R,10aS,12R,14aR)-2,9-bis(6-amino-9H-purin-9-yl)octahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8 ]tetraoxadiphosphacyclododecine-3,5,10,12-tetrol 5,12-dioxide, SULFATE ION, Stimulator of interferon genes protein | | 著者 | Yuan, Z.L, Shang, G.J, Cong, X.Y, Gu, L.C. | | 登録日 | 2018-12-15 | | 公開日 | 2019-06-19 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (1.764 Å) | | 主引用文献 | Crystal structures of porcine STINGCBD-CDN complexes reveal the mechanism of ligand recognition and discrimination of STING proteins.
J.Biol.Chem., 294, 2019
|
|
5ZO3
 
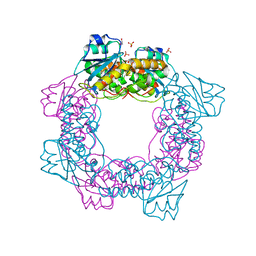 | | apo form of the nuclease | | 分子名称: | 1,2-ETHANEDIOL, Putative 3'-5' exonuclease family protein, SULFATE ION | | 著者 | Yuan, Z.L, Gu, L.C. | | 登録日 | 2018-04-12 | | 公開日 | 2019-04-10 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (1.493 Å) | | 主引用文献 | NrnC, an RNase D-Like Protein FromAgrobacterium, Is a Novel Octameric Nuclease That Specifically Degrades dsDNA but Leaves dsRNA Intact.
Front Microbiol, 9, 2018
|
|
5ZO4
 
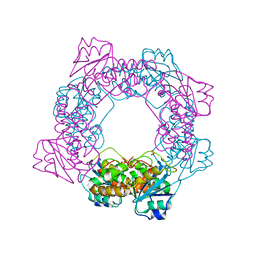 | | inactive state of the nuclease | | 分子名称: | MANGANESE (II) ION, Putative 3'-5' exonuclease family protein, SULFATE ION | | 著者 | Yuan, Z.L, Gu, L.C. | | 登録日 | 2018-04-12 | | 公開日 | 2019-04-10 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | NrnC, an RNase D-Like Protein FromAgrobacterium, Is a Novel Octameric Nuclease That Specifically Degrades dsDNA but Leaves dsRNA Intact.
Front Microbiol, 9, 2018
|
|
5ZO5
 
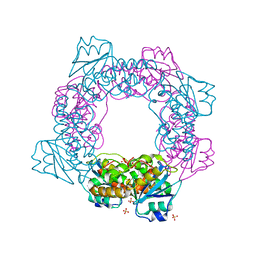 | | active state of the nuclease | | 分子名称: | MANGANESE (II) ION, Putative 3'-5' exonuclease family protein, SULFATE ION | | 著者 | Yuan, Z.L, Gu, L.C. | | 登録日 | 2018-04-12 | | 公開日 | 2019-04-10 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (2.297 Å) | | 主引用文献 | NrnC, an RNase D-Like Protein FromAgrobacterium, Is a Novel Octameric Nuclease That Specifically Degrades dsDNA but Leaves dsRNA Intact.
Front Microbiol, 9, 2018
|
|
6A06
 
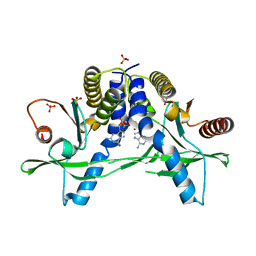 | | Structure of pSTING complex | | 分子名称: | SULFATE ION, Stimulator of interferon genes protein, cGAMP | | 著者 | Yuan, Z.L, Shang, G.J, Cong, X.Y, Gu, L.C. | | 登録日 | 2018-06-05 | | 公開日 | 2019-06-19 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (1.792 Å) | | 主引用文献 | Crystal structures of porcine STINGCBD-CDN complexes reveal the mechanism of ligand recognition and discrimination of STING proteins.
J.Biol.Chem., 294, 2019
|
|
6A04
 
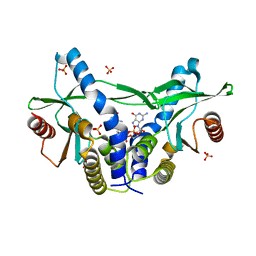 | | Structure of pSTING complex | | 分子名称: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), SULFATE ION, Stimulator of interferon genes protein | | 著者 | Yuan, Z.L, Shang, G.J, Cong, X.Y, Gu, L.C. | | 登録日 | 2018-06-05 | | 公開日 | 2019-06-19 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Crystal structures of porcine STINGCBD-CDN complexes reveal the mechanism of ligand recognition and discrimination of STING proteins.
J.Biol.Chem., 294, 2019
|
|
6A05
 
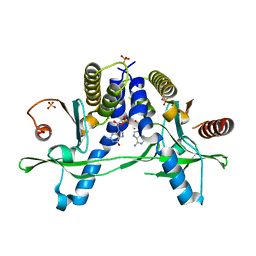 | | Structure of pSTING complex | | 分子名称: | 2-amino-9-[(2R,3R,3aR,5S,7aS,9R,10R,10aR,12R,14aS)-9-(6-amino-9H-purin-9-yl)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecin-2-yl]-1,9-dihydro-6H-purin-6-one, SULFATE ION, Stimulator of interferon genes protein | | 著者 | Yuan, Z.L, Shang, G.J, Cong, X.Y, Gu, L.C. | | 登録日 | 2018-06-05 | | 公開日 | 2019-06-19 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Crystal structures of porcine STINGCBD-CDN complexes reveal the mechanism of ligand recognition and discrimination of STING proteins.
J.Biol.Chem., 294, 2019
|
|
