2EHG
 
 | |
4E19
 
 | |
4NYN
 
 | |
2Z1I
 
 | | Crystal structure of E.coli RNase HI surface charged mutant(Q4R/T40E/Q72H/Q76K/Q80E/T92K/Q105K/Q113R/Q115K) | | Descriptor: | Ribonuclease HI | | Authors: | You, D.J, Fukuchi, S, Nishikawa, K, Koga, Y, Takano, K, Kanaya, S. | | Deposit date: | 2007-05-08 | | Release date: | 2007-11-13 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Protein Thermostabilization Requires a Fine-tuned Placement of Surface-charged Residues
J.Biochem.(Tokyo), 142, 2007
|
|
2Z1J
 
 | | Crystal structure of E.coli RNase HI surface charged mutant(Q4R/T40E/Q72H/Q76K/Q80E/T92K/Q105K/Q113R/Q115K/N143K/T145K) | | Descriptor: | Ribonuclease HI | | Authors: | You, D.J, Fukuchi, S, Nishikawa, K, Koga, Y, Takano, K, Kanaya, S. | | Deposit date: | 2007-05-08 | | Release date: | 2007-11-13 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Protein Thermostabilization Requires a Fine-tuned Placement of Surface-charged Residues
J.Biochem.(Tokyo), 142, 2007
|
|
2Z1H
 
 | | Crystal structure of E.coli RNase HI surface charged mutant(Q4R/T92K/Q105K/Q113R/Q115K/N143K/T145K) | | Descriptor: | Ribonuclease HI | | Authors: | You, D.J, Fukuchi, S, Nishikawa, K, Koga, Y, Takano, K, Kanaya, S. | | Deposit date: | 2007-05-08 | | Release date: | 2007-11-13 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Protein Thermostabilization Requires a Fine-tuned Placement of Surface-charged Residues
J.Biochem.(Tokyo), 142, 2007
|
|
2Z1G
 
 | | Crystal structure of E.coli RNase HI surface charged mutant(Q4R/T40E/Q72H/Q76K/Q80E/T92K/Q105K) | | Descriptor: | Ribonuclease HI | | Authors: | You, D.J, Fukuchi, S, Nishikawa, K, Koga, Y, Takano, K, Kanaya, S. | | Deposit date: | 2007-05-08 | | Release date: | 2007-11-13 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Protein Thermostabilization Requires a Fine-tuned Placement of Surface-charged Residues
J.Biochem.(Tokyo), 142, 2007
|
|
3VV2
 
 | | Crystal structure of complex form between S324A-subtilisin and mutant Tkpro | | Descriptor: | CALCIUM ION, CHLORIDE ION, PROPEPTIDE from Tk-subtilisin, ... | | Authors: | Uehara, R, Ueda, Y, You, D.J, Takano, K, Koga, Y, Kanaya, S. | | Deposit date: | 2012-07-12 | | Release date: | 2013-03-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Accelerated maturation of Tk-subtilisin by a Leu Pro mutation at the C-terminus of the propeptide, which reduces the binding of the propeptide to Tk-subtilisin
Febs J., 280, 2013
|
|
3VN5
 
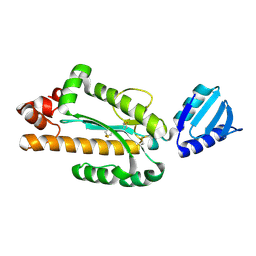 | | Crystal structure of Aquifex aeolicus RNase H3 | | Descriptor: | Ribonuclease HIII | | Authors: | Jongruja, N, You, D.J, Eiko, K, Angkawidjaja, C, Koga, Y, Takano, K, Kanaya, S. | | Deposit date: | 2011-12-22 | | Release date: | 2012-12-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structure and characterization of RNase H3 from Aquifex aeolicus
Febs J., 279, 2012
|
|
4JP8
 
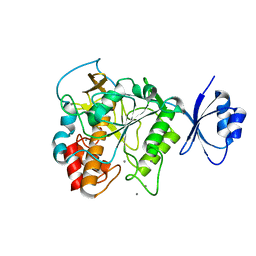 | | Crystal structure of Pro-F17H/S324A | | Descriptor: | CALCIUM ION, Tk-subtilisin | | Authors: | Yuzaki, K, You, D.J, Uehara, R, Koga, Y, Kanaya, S. | | Deposit date: | 2013-03-19 | | Release date: | 2014-01-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Increase in activation rate of Pro-Tk-subtilisin by a single nonpolar-to-polar amino acid substitution at the hydrophobic core of the propeptide domain
Protein Sci., 22, 2013
|
|
4MAD
 
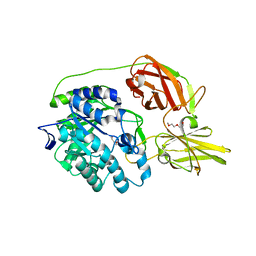 | | Crystal structure of beta-galactosidase C (BgaC) from Bacillus circulans ATCC 31382 | | Descriptor: | 2-(2-METHOXYETHOXY)ETHANOL, Beta-galactosidase | | Authors: | Kamerke, C, You, D.J, Kanaya, S, Elling, L. | | Deposit date: | 2013-08-16 | | Release date: | 2014-08-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Rational design of a glycosynthase by the crystal structure of beta-galactosidase from Bacillus circulans (BgaC) and its use for the synthesis of N-acetyllactosamine type 1 glycan structures.
J.Biotechnol., 191, 2014
|
|
2E4L
 
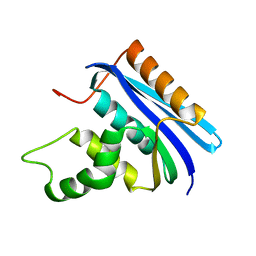 | | Thermodynamic and Structural Analysis of Thermolabile RNase HI from Shewanella oneidensis MR-1 | | Descriptor: | Ribonuclease HI | | Authors: | Tadokoro, T, You, D.J, Chon, H, Matsumura, H, Koga, Y, Takano, K, Kanaya, S. | | Deposit date: | 2006-12-13 | | Release date: | 2007-05-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural, thermodynamic, and mutational analyses of a psychrotrophic RNase HI.
Biochemistry, 46, 2007
|
|
4EB0
 
 | | Crystal structure of Leaf-branch compost bacterial cutinase homolog | | Descriptor: | LCC, THIOCYANATE ION | | Authors: | Sulaiman, S, You, D.J, Eiko, K, Koga, Y, Kanaya, S. | | Deposit date: | 2012-03-23 | | Release date: | 2013-03-27 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of Leaf-branch compost bacterial cutinase homolog
To be Published
|
|
4H8K
 
 | | Crystal structure of LC11-RNase H1 in complex with RNA/DNA hybrid | | Descriptor: | DNA (5'-D(*GP*GP*AP*AP*TP*CP*AP*GP*GP*TP*GP*TP*CP*G)-3'), RNA (5'-R(*CP*GP*AP*CP*AP*CP*CP*UP*GP*AP*UP*UP*CP*C)-3'), Ribonuclease H | | Authors: | Nguyen, T.N, You, D.J, Matsumoto, H, Kanaya, E, Kanaya, S. | | Deposit date: | 2012-09-23 | | Release date: | 2013-09-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of metagenome-derived LC11-RNase H1 in complex with RNA/DNA hybrid
J.Struct.Biol., 182, 2013
|
|
4OEC
 
 | | Crystal structure of glycerophosphodiester phosphodiesterase from Thermococcus kodakarensis KOD1 | | Descriptor: | Glycerophosphoryl diester phosphodiesterase, MAGNESIUM ION | | Authors: | Atsuta, Y, You, D.J, Takano, K, Koga, Y, Kanaya, S. | | Deposit date: | 2014-01-13 | | Release date: | 2015-01-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of glycerophosphodiester phosphodiesterase from Thermococcus kodakarensis KOD1
To be Published
|
|
2Z57
 
 | | Crystal structure of G56E-propeptide:S324A-subtilisin complex | | Descriptor: | CALCIUM ION, Tk-subtilisin, ZINC ION | | Authors: | Pulido, M.A, Tanaka, S, Sringiew, C, You, D.J, Matsumura, H, Koga, Y, Takano, K, Kanaya, S. | | Deposit date: | 2007-06-29 | | Release date: | 2008-01-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Requirement of left-handed glycine residue for high stability of the Tk-subtilisin propeptide as revealed by mutational and crystallographic analyses
J.Mol.Biol., 374, 2007
|
|
2Z56
 
 | | Crystal structure of G56S-propeptide:S324A-subtilisin complex | | Descriptor: | CALCIUM ION, Tk-subtilisin, ZINC ION | | Authors: | Pulido, M.A, Tanaka, S, Sringiew, C, You, D.J, Matsumura, H, Koga, Y, Takano, K, Kanaya, S. | | Deposit date: | 2007-06-29 | | Release date: | 2008-01-01 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Requirement of left-handed glycine residue for high stability of the Tk-subtilisin propeptide as revealed by mutational and crystallographic analyses
J.Mol.Biol., 374, 2007
|
|
2Z8Z
 
 | | Crystal structure of a platinum-bound S445C mutant of Pseudomonas sp. MIS38 lipase | | Descriptor: | CALCIUM ION, Lipase, PLATINUM (II) ION, ... | | Authors: | Angkawidjaja, C, You, D.J, Matsumura, H, Koga, Y, Takano, K, Kanaya, S. | | Deposit date: | 2007-09-13 | | Release date: | 2007-10-30 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of a family I.3 lipase from Pseudomonas sp. MIS38 in a closed conformation
FEBS Lett., 581, 2007
|
|
2Z58
 
 | | Crystal structure of G56W-propeptide:S324A-subtilisin complex | | Descriptor: | CALCIUM ION, Tk-subtilisin, ZINC ION | | Authors: | Pulido, M.A, Tanaka, S, Sringiew, C, You, D.J, Matsumura, H, Koga, Y, Takano, K, Kanaya, S. | | Deposit date: | 2007-06-29 | | Release date: | 2008-01-01 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Requirement of left-handed glycine residue for high stability of the Tk-subtilisin propeptide as revealed by mutational and crystallographic analyses
J.Mol.Biol., 374, 2007
|
|
2Z8X
 
 | | Crystal structure of extracellular lipase from Pseudomonas sp. MIS38 | | Descriptor: | CALCIUM ION, Lipase, ZINC ION | | Authors: | Angkawidjaja, C, You, D.J, Matsumura, H, Koga, Y, Takano, K, Kanaya, S. | | Deposit date: | 2007-09-11 | | Release date: | 2007-10-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Crystal structure of a family I.3 lipase from Pseudomonas sp. MIS38 in a closed conformation
FEBS Lett., 581, 2007
|
|
