7CBV
 
 | |
7E90
 
 | |
7E92
 
 | |
7CUS
 
 | |
7F2H
 
 | |
7F2G
 
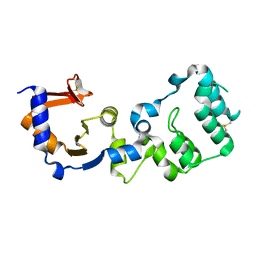 | |
7C7Z
 
 | |
6IWY
 
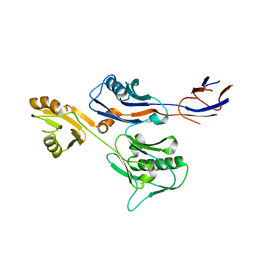 | |
6KNT
 
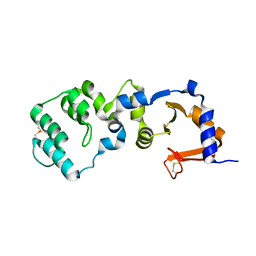
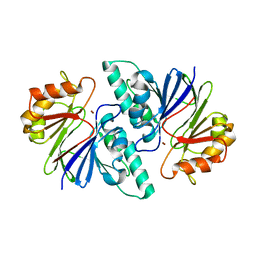 | | Crystal structure of the metallo-beta-lactamase fold protein YhfI from Bacillus subtilis (space group P4332) | | Descriptor: | Putative metal-dependent hydrolase, ZINC ION | | Authors: | Na, H.W, Namgung, B, Song, W.S, Yoon, S.I. | | Deposit date: | 2019-08-07 | | Release date: | 2019-09-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural and biochemical analyses of the metallo-beta-lactamase fold protein YhfI from Bacillus subtilis.
Biochem.Biophys.Res.Commun., 519, 2019
|
|
6JV6
 
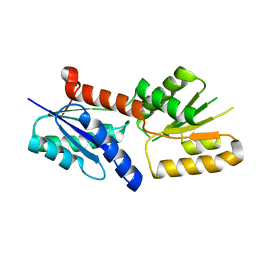 | | Crystal structure of the sirohydrochlorin chelatase SirB from Bacillus subtilis subspecies spizizenii in complex with cobalt | | Descriptor: | COBALT (II) ION, Sirohydrochlorin ferrochelatase | | Authors: | Nam, M.S, Song, W.S, Park, S.C, Yoon, S.I. | | Deposit date: | 2019-04-16 | | Release date: | 2019-06-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Cobalt complex structure of the sirohydrochlorin chelatase SirB from Bacillus subtilis subsp. spizizenii.
KOREAN J MICROBIOL., 55, 2019
|
|
6JYI
 
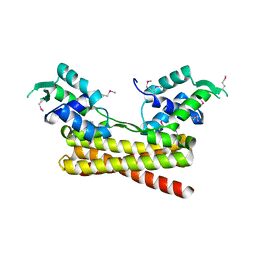 | | Crystal structure of the PadR-like transcriptional regulator BC1756 from Bacillus cereus | | Descriptor: | Transcriptional repressor PadR | | Authors: | Kim, T.H, Park, S.C, Lee, K.C, Song, W.S, Yoon, S.I. | | Deposit date: | 2019-04-26 | | Release date: | 2019-06-26 | | Last modified: | 2019-07-10 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Structural and DNA-binding studies of the PadR-like transcriptional regulator BC1756 from Bacillus cereus.
Biochem.Biophys.Res.Commun., 515, 2019
|
|
6KNS
 
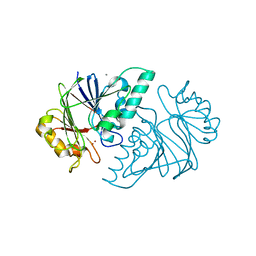 | | Crystal structure of the metallo-beta-lactamase fold protein YhfI from Bacillus subtilis (space group I4122) | | Descriptor: | CALCIUM ION, Putative metal-dependent hydrolase, ZINC ION | | Authors: | Na, H.W, Namgung, B, Song, W.S, Yoon, S.I. | | Deposit date: | 2019-08-07 | | Release date: | 2019-09-18 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural and biochemical analyses of the metallo-beta-lactamase fold protein YhfI from Bacillus subtilis.
Biochem.Biophys.Res.Commun., 519, 2019
|
|
6KTY
 
 | |
7X1K
 
 | |
7XFP
 
 | |
7W1F
 
 | | Crystal structure of the dNTP triphosphohydrolase PA1124 from Pseudomonas aeruginosa | | Descriptor: | NICKEL (II) ION, Probable deoxyguanosinetriphosphate triphosphohydrolase | | Authors: | Oh, H.B, Song, W.S, Lee, K.C, Park, S.C, Yoon, S.I. | | Deposit date: | 2021-11-19 | | Release date: | 2022-03-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural analysis of the dNTP triphosphohydrolase PA1124 from Pseudomonas aeruginosa.
Biochem.Biophys.Res.Commun., 589, 2022
|
|
7X9R
 
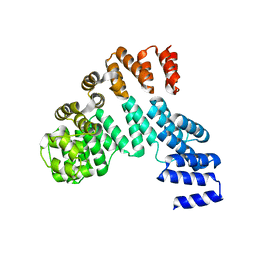 | | Crystal structure of the antirepressor GmaR | | Descriptor: | Glycosyl transferase family 2 | | Authors: | Cho, S.Y, Na, H.W, Oh, H.B, Kwak, Y.M, Song, W.S, Park, S.C, Yoon, S.I. | | Deposit date: | 2022-03-16 | | Release date: | 2022-11-09 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural basis of flagellar motility regulation by the MogR repressor and the GmaR antirepressor in Listeria monocytogenes.
Nucleic Acids Res., 50, 2022
|
|
7X9S
 
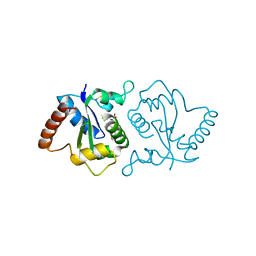
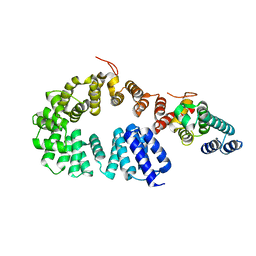 | | Crystal structure of a complex between the antirepressor GmaR and the transcriptional repressor MogR | | Descriptor: | GmaR, Motility gene repressor MogR | | Authors: | Cho, S.Y, Na, H.W, Oh, H.B, Kwak, Y.M, Song, W.S, Park, S.C, Yoon, S.I. | | Deposit date: | 2022-03-16 | | Release date: | 2022-11-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.11 Å) | | Cite: | Structural basis of flagellar motility regulation by the MogR repressor and the GmaR antirepressor in Listeria monocytogenes.
Nucleic Acids Res., 50, 2022
|
|
7YLG
 
 | |
7YLF
 
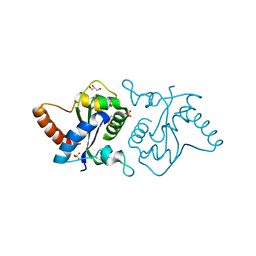 | |
5YHH
 
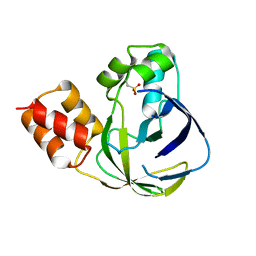 | | Crystal structure of YiiM from Geobacillus stearothermophilus | | Descriptor: | Uncharacterized conserved protein YiiM | | Authors: | Namgung, B, Kim, J.H, Song, W.S, Yoon, S.I. | | Deposit date: | 2017-09-28 | | Release date: | 2018-03-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the hydroxylaminopurine resistance protein, YiiM, and its putative molybdenum cofactor-binding catalytic site.
Sci Rep, 8, 2018
|
|
5Y8T
 
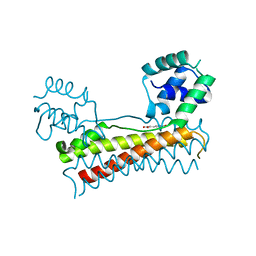 | | Crystal structure of Bacillus subtilis PadR in complex with p-coumaric acid | | Descriptor: | 4'-HYDROXYCINNAMIC ACID, Transcriptional regulator | | Authors: | Park, S.C, Kwak, Y.M, Song, W.S, Hong, M, Yoon, S.I. | | Deposit date: | 2017-08-21 | | Release date: | 2017-11-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis of effector and operator recognition by the phenolic acid-responsive transcriptional regulator PadR.
Nucleic Acids Res., 45, 2017
|
|
5YHI
 
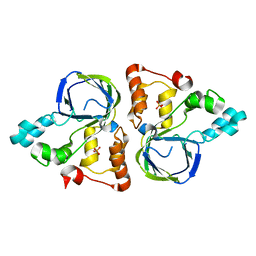 | | Crystal structure of YiiM from Escherichia coli | | Descriptor: | PHOSPHATE ION, Protein YiiM | | Authors: | Namgung, B, Kim, J.H, Song, W.S, Yoon, S.I. | | Deposit date: | 2017-09-28 | | Release date: | 2018-03-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Crystal structure of the hydroxylaminopurine resistance protein, YiiM, and its putative molybdenum cofactor-binding catalytic site.
Sci Rep, 8, 2018
|
|
5ZIY
 
 | | Crystal structure of Bacillus cereus FlgL | | Descriptor: | Flagellar hook-associated protein 3, ZINC ION | | Authors: | Hong, H.J, Kim, T.H, Song, W.S, Yoon, S.I. | | Deposit date: | 2018-03-18 | | Release date: | 2018-10-17 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of FlgL and its implications for flagellar assembly
Sci Rep, 8, 2018
|
|
5ZJ0
 
 | |
