8XJE
 
 | |
8YJX
 
 | |
8ZAH
 
 | |
8ZAK
 
 | |
4XPK
 
 | | The crystal structure of Campylobacter jejuni N-acetyltransferase PseH | | Descriptor: | N-Acetyltransferase, PseH | | Authors: | Song, W.S, Nam, M.S, Namgung, B, Yoon, S.I. | | Deposit date: | 2015-01-17 | | Release date: | 2015-03-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural analysis of PseH, the Campylobacter jejuni N-acetyltransferase involved in bacterial O-linked glycosylation.
Biochem.Biophys.Res.Commun., 458, 2015
|
|
4XPL
 
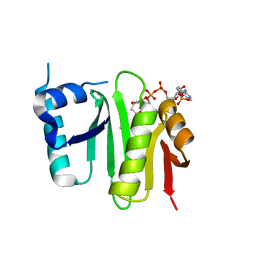 | | The crystal structure of Campylobacter jejuni N-acetyltransferase PseH in complex with acetyl coenzyme A | | Descriptor: | ACETYL COENZYME *A, N-Acetyltransferase, PseH | | Authors: | Song, W.S, Nam, M.S, Namgung, B, Yoon, S.I. | | Deposit date: | 2015-01-17 | | Release date: | 2015-03-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural analysis of PseH, the Campylobacter jejuni N-acetyltransferase involved in bacterial O-linked glycosylation.
Biochem.Biophys.Res.Commun., 458, 2015
|
|
8H50
 
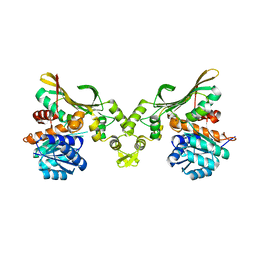 | |
8H52
 
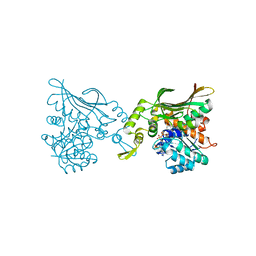 | | Crystal structure of Helicobacter pylori carboxyspermidine dehydrogenase in complex with NADP | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Saccharopine dehydrogenase | | Authors: | Ko, K.Y, Park, S.C, Cho, S.Y, Yoon, S.I. | | Deposit date: | 2022-10-11 | | Release date: | 2022-11-09 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural analysis of carboxyspermidine dehydrogenase from Helicobacter pylori.
Biochem.Biophys.Res.Commun., 635, 2022
|
|
8H4Z
 
 | |
8IEU
 
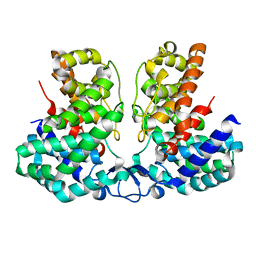 | |
8IEV
 
 | |
8J56
 
 | | Crystal structure of the FlhDC complex from Cupriavidus necator | | Descriptor: | Flagellar transcriptional regulator FlhC, Flagellar transcriptional regulator FlhD, ZINC ION | | Authors: | Cho, S.Y, Oh, H.B, Yoon, S.I. | | Deposit date: | 2023-04-21 | | Release date: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Hexameric structure of the flagellar master regulator FlhDC from Cupriavidus necator and its interaction with flagellar promoter DNA.
Biochem.Biophys.Res.Commun., 672, 2023
|
|
5X12
 
 | | Crystal structure of Bacillus subtilis PadR | | Descriptor: | Transcriptional regulator | | Authors: | Park, S.C, Kwak, Y.M, Song, W.S, Hong, M, Yoon, S.I. | | Deposit date: | 2017-01-24 | | Release date: | 2017-11-22 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural basis of effector and operator recognition by the phenolic acid-responsive transcriptional regulator PadR
Nucleic Acids Res., 45, 2017
|
|
5X11
 
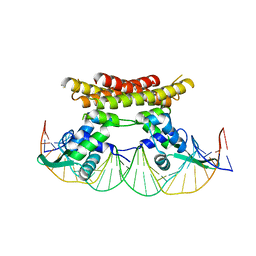 | | Crystal structure of Bacillus subtilis PadR in complex with operator DNA | | Descriptor: | DNA (28-MER), Transcriptional regulator | | Authors: | Park, S.C, Kwak, Y.M, Song, W.S, Hong, M, Yoon, S.I. | | Deposit date: | 2017-01-24 | | Release date: | 2017-11-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structural basis of effector and operator recognition by the phenolic acid-responsive transcriptional regulator PadR
Nucleic Acids Res., 45, 2017
|
|
5X14
 
 | | Crystal structure of Bacillus subtilis PadR in complex with ferulic acid | | Descriptor: | 3-(4-HYDROXY-3-METHOXYPHENYL)-2-PROPENOIC ACID, GLYCEROL, Transcriptional regulator | | Authors: | Park, S.C, Kwak, Y.M, Song, W.S, Hong, M, Yoon, S.I. | | Deposit date: | 2017-01-24 | | Release date: | 2017-11-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Structural basis of effector and operator recognition by the phenolic acid-responsive transcriptional regulator PadR
Nucleic Acids Res., 45, 2017
|
|
5X13
 
 | | Crystal structure of Bacillus subtilis PadR in complex with p-coumaric acid | | Descriptor: | 4'-HYDROXYCINNAMIC ACID, GLYCEROL, Transcriptional regulator | | Authors: | Park, S.C, Kwak, Y.M, Song, W.S, Hong, M, Yoon, S.I. | | Deposit date: | 2017-01-24 | | Release date: | 2017-11-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural basis of effector and operator recognition by the phenolic acid-responsive transcriptional regulator PadR
Nucleic Acids Res., 45, 2017
|
|
5DQV
 
 | | The crystal structure of Bacillus subtilis YpgQ | | Descriptor: | NICKEL (II) ION, Uncharacterized protein | | Authors: | Jeon, Y.J, Song, W.S, Yoon, S.I. | | Deposit date: | 2015-09-15 | | Release date: | 2016-04-27 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and biochemical characterization of bacterial YpgQ protein reveals a metal-dependent nucleotide pyrophosphohydrolase
J.Struct.Biol., 195, 2016
|
|
5DQW
 
 | |
5H5V
 
 | |
5H5W
 
 | |
5H5T
 
 | |
5IHY
 
 | |
5GY2
 
 | |
3UN9
 
 | | Crystal structure of an immune receptor | | Descriptor: | NLR family member X1, PLATINUM (II) ION | | Authors: | Hong, M, Yoon, S.I, Wilson, I.A. | | Deposit date: | 2011-11-15 | | Release date: | 2012-03-28 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structure and Functional Characterization of the RNA-Binding Element of the NLRX1 Innate Immune Modulator.
Immunity, 36, 2012
|
|
8H2C
 
 | | Crystal structure of the pseudaminic acid synthase PseI from Campylobacter jejuni | | Descriptor: | MANGANESE (II) ION, Pseudaminic acid synthase | | Authors: | Song, W.S, Park, M.A, Ki, D.U, Yoon, S.I. | | Deposit date: | 2022-10-05 | | Release date: | 2022-11-09 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural analysis of the pseudaminic acid synthase PseI from Campylobacter jejuni.
Biochem.Biophys.Res.Commun., 635, 2022
|
|
