4W7F
 
 | |
2Z9H
 
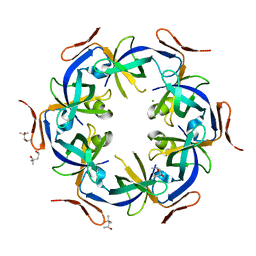 | | Ethanolamine utilization protein, EutN | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, CHLORIDE ION, Ethanolamine utilization protein eutN | | Authors: | Tanaka, S, Sawaya, M.R, Yeates, T.O. | | Deposit date: | 2007-09-20 | | Release date: | 2007-10-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | The crystal structure of ethanolamine utilization protein EutN from E. coli
To be Published
|
|
3BN4
 
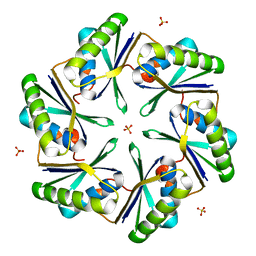 | | Carboxysome Subunit, CcmK1 | | Descriptor: | Carbon dioxide-concentrating mechanism protein ccmK homolog 1, SULFATE ION | | Authors: | Tanaka, S, Sawaya, M.R, Yeates, T.O. | | Deposit date: | 2007-12-13 | | Release date: | 2008-03-04 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Atomic-level models of the bacterial carboxysome shell.
Science, 319, 2008
|
|
5UI2
 
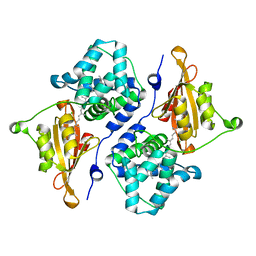 | | CRYSTAL STRUCTURE OF ORANGE CAROTENOID PROTEIN | | Descriptor: | (3'R)-3'-hydroxy-beta,beta-caroten-4-one, CHLORIDE ION, Orange carotenoid-binding protein, ... | | Authors: | KERFELD, C.A, SAWAYA, M.R, VISHNU, B, KROGMANN, D, YEATES, T.O. | | Deposit date: | 2017-01-12 | | Release date: | 2017-01-25 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The crystal structure of a cyanobacterial water-soluble carotenoid binding protein.
Structure, 11, 2003
|
|
1YAC
 
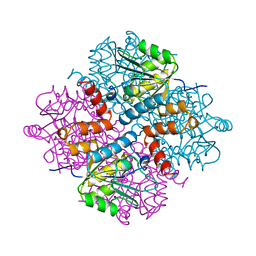 | |
2B5O
 
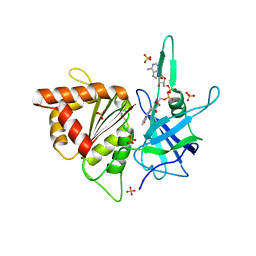 | | ferredoxin-NADP reductase | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Ferredoxin--NADP reductase, SULFATE ION | | Authors: | Sawaya, M.R, Kerfeld, C.A, Gomez-Lojero, C, Krogmann, D, Bryant, D.A, Yeates, T.O. | | Deposit date: | 2005-09-29 | | Release date: | 2005-10-11 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.499 Å) | | Cite: | Crystal Structure of Ferredoxin-NADP reductase from Synechococcus sp. (PCC 7002)
To be Published
|
|
2EWH
 
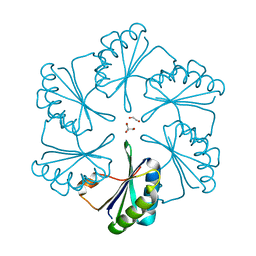 | | Carboxysome protein CsoS1A from Halothiobacillus neapolitanus | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Major carboxysome shell protein 1A | | Authors: | Tsai, Y, Sawaya, M.R, Kerfeld, C.A, Yeates, T.O. | | Deposit date: | 2005-11-03 | | Release date: | 2006-11-14 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural Analysis of CsoS1A and the Protein Shell of the Halothiobacillus neapolitanus Carboxysome.
Plos Biol., 5, 2007
|
|
2Q31
 
 | | Actin Dimer Cross-linked Between Residues 41 and 374 and proteolytically cleaved by subtilisin between residues 47 and 48. | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Sawaya, M.R, Pashkov, I, Kudryashov, D.S, Reisler, E, Yeates, T.O. | | Deposit date: | 2007-05-29 | | Release date: | 2007-06-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Multiple crystal structures of actin dimers and their implications for interactions in the actin filament.
Acta Crystallogr.,Sect.D, 64, 2008
|
|
6C9K
 
 | | Single-Particle reconstruction of DARP14 - A designed protein scaffold displaying ~17kDa DARPin proteins | | Descriptor: | DARP14 - Subunit A with DARPin, DARP14 - Subunit B | | Authors: | Gonen, S, Liu, Y, Yeates, T.O, Gonen, T. | | Deposit date: | 2018-01-26 | | Release date: | 2018-03-21 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.49 Å) | | Cite: | Near-atomic cryo-EM imaging of a small protein displayed on a designed scaffolding system.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6C9I
 
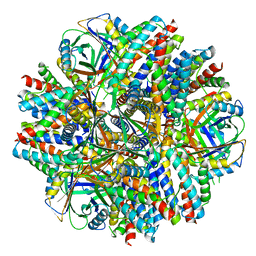 | | Single-Particle reconstruction of DARP14 - A designed protein scaffold displaying ~17kDa DARPin proteins - Scaffold | | Descriptor: | DARP14 - Subunit A with DARPin, DARP14 - Subunit B | | Authors: | Gonen, S, Liu, Y, Yeates, T.O, Gonen, T. | | Deposit date: | 2018-01-26 | | Release date: | 2018-03-21 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.09 Å) | | Cite: | Near-atomic cryo-EM imaging of a small protein displayed on a designed scaffolding system.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
2PLT
 
 | |
2Q1N
 
 | | Actin Dimer Cross-linked Between Residues 41 and 374 | | Descriptor: | Actin, alpha skeletal muscle, CALCIUM ION, ... | | Authors: | Sawaya, M.R, Pashkov, I, Kudryashov, D.S, Adisetiyo, H, Reisler, E, Yeates, T.O. | | Deposit date: | 2007-05-25 | | Release date: | 2007-06-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Multiple crystal structures of actin dimers and their implications for interactions in the actin filament.
Acta Crystallogr.,Sect.D, 64, 2008
|
|
2Q36
 
 | | Actin Dimer Cross-linked between Residues 191 and 374 and complexed with Kabiramide C | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Sawaya, M.R, Pashkov, I, Kudryashov, D.S, Reisler, E, Yeates, T.O. | | Deposit date: | 2007-05-29 | | Release date: | 2007-06-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Multiple crystal structures of actin dimers and their implications for interactions in the actin filament.
Acta Crystallogr.,Sect.D, 64, 2008
|
|
6ARD
 
 | |
6ARC
 
 | |
1C3C
 
 | | T. MARITIMA ADENYLOSUCCINATE LYASE | | Descriptor: | PROTEIN (ADENYLOSUCCINATE LYASE) | | Authors: | Toth, E.A, Yeates, T.O. | | Deposit date: | 1999-07-27 | | Release date: | 2000-02-09 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The structure of adenylosuccinate lyase, an enzyme with dual activity in the de novo purine biosynthetic pathway.
Structure Fold.Des., 8, 2000
|
|
1C3U
 
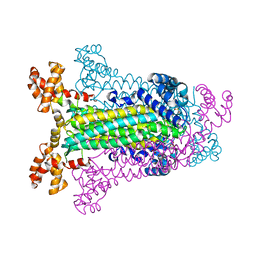 | | T. MARITIMA ADENYLOSUCCINATE LYASE | | Descriptor: | ADENYLOSUCCINATE LYASE, SULFATE ION | | Authors: | Toth, E.A, Yeates, T.O. | | Deposit date: | 1999-07-28 | | Release date: | 2000-03-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The structure of adenylosuccinate lyase, an enzyme with dual activity in the de novo purine biosynthetic pathway.
Structure Fold.Des., 8, 2000
|
|
2A1B
 
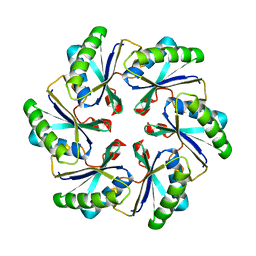 | | Carboxysome shell protein ccmK2 | | Descriptor: | Carbon dioxide concentrating mechanism protein ccmK homolog 2 | | Authors: | Kerfeld, C.A, Sawaya, M.R, Tanaka, S, Nguyen, C.V, Phillips, M, Beeby, M, Yeates, T.O. | | Deposit date: | 2005-06-20 | | Release date: | 2005-08-09 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Protein structures forming the shell of primitive bacterial organelles
Science, 309, 2005
|
|
2A10
 
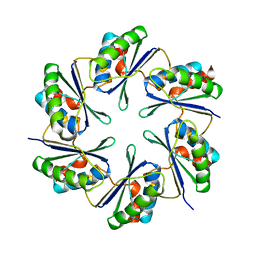 | | carboxysome shell protein ccmK4 | | Descriptor: | Carbon dioxide concentrating mechanism protein ccmK homolog 4 | | Authors: | Kerfeld, C.A, Sawaya, M.R, Tanaka, S, Nguyen, C.V, Phillips, M, Beeby, M, Yeates, T.O. | | Deposit date: | 2005-06-17 | | Release date: | 2005-08-09 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.803 Å) | | Cite: | Protein structures forming the shell of primitive bacterial organelles
Science, 309, 2005
|
|
2A5X
 
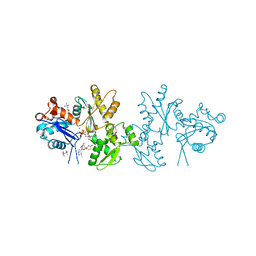 | | Crystal Structure of a Cross-linked Actin Dimer | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Actin, alpha skeletal muscle, ... | | Authors: | Kudryashov, D.S, Sawaya, M.R, Adisetiyo, H, Norcross, T, Hegyi, G, Reisler, E, Yeates, T.O. | | Deposit date: | 2005-07-01 | | Release date: | 2005-08-23 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | The crystal structure of a cross-linked actin dimer suggests a detailed molecular interface in F-actin
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
2A18
 
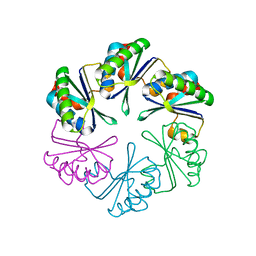 | | carboxysome shell protein ccmK4, crystal form 2 | | Descriptor: | AMMONIUM ION, Carbon dioxide concentrating mechanism protein ccmK homolog 4 | | Authors: | Kerfeld, C.A, Sawaya, M.R, Tanaka, S, Nguyen, C.V, Phillips, M, Beeby, M, Yeates, T.O. | | Deposit date: | 2005-06-18 | | Release date: | 2005-08-09 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Protein structures forming the shell of primitive bacterial organelles
Science, 309, 2005
|
|
4D9J
 
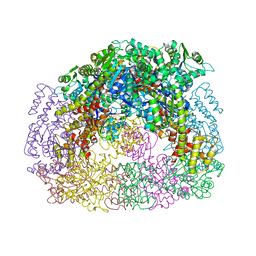 | |
4DDF
 
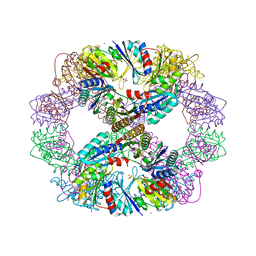 | | Computationally Designed Self-assembling Octahedral Cage protein, O333, Crystallized in space group P4 | | Descriptor: | CHLORIDE ION, Propanediol utilization polyhedral body protein PduT, SULFATE ION | | Authors: | Sawaya, M.R, King, N.P, Sheffler, W, Baker, D, Yeates, T.O. | | Deposit date: | 2012-01-18 | | Release date: | 2012-06-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Computational design of self-assembling protein nanomaterials with atomic level accuracy.
Science, 336, 2012
|
|
4DCL
 
 | | Computationally Designed Self-assembling tetrahedron protein, T308, Crystallized in space group F23 | | Descriptor: | Putative acetyltransferase SACOL2570 | | Authors: | Sawaya, M.R, King, N.P, Sheffler, W, Baker, D, Yeates, T.O. | | Deposit date: | 2012-01-17 | | Release date: | 2012-06-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | Computational design of self-assembling protein nanomaterials with atomic level accuracy.
Science, 336, 2012
|
|
2PY8
 
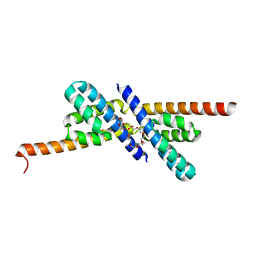 | | RbcX | | Descriptor: | 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, CHLORIDE ION, Hypothetical protein rbcX | | Authors: | Tanaka, S, Sawaya, M.R, Kerfeld, C.A, Yeates, T.O. | | Deposit date: | 2007-05-15 | | Release date: | 2007-10-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structure of the RuBisCO chaperone RbcX from Synechocystis sp. PCC6803.
Acta Crystallogr.,Sect.D, 63, 2007
|
|
